MCB TRANSCRIPT

Translational Regulation: How Cells Control Protein Synthesis
When MCB Assistant Professor Nicholas Ingolia was a post-doc, he developed a long-sought technique — called ribosome profiling — that gives a snapshot of mRNA translation into proteins.

“You immobilize the ribosomes, use nucleases to digest the exposed mRNA, and sequence the protected areas,” he explains. “It’s a great way to measure what and how much is being translated.” The ribosome-protected mRNA footprints are long enough to be matched to specific genes in most cases.
Now, Ingolia uses ribosome profiling to find out how cells regulate mRNA translation, which is thought to affect protein levels as much as the transcription of genes into mRNAs. In people, for example, there’s a 20 to 30-fold difference in the number of protein molecules made from a given mRNA. “Protein synthesis is the true end point of gene expression,” he says.
His lab is asking three main questions about translational control of gene expression, using yeast and human cells. One is how do cells control where translation starts? This wasn’t even in question until recently because translation had generally been thought to start at AUG codons. But ribosomal profiling showed otherwise. “More than half of potential starting places are unconventional,” Ingolia says.
These alternate start sites can affect how much of a protein is synthesized, and can also lead to production of forms of a protein with different functions. “We believe start codon selection is an underappreciated control point for gene expression,” Ingolia says.
Another question is how do drugs stop translation of some mRNAs but not others? Take rocaglamide, a plant-derived compound used in traditional Chinese medicine that kills cancer cells preferentially. “Rocaglamide targets an RNA helicase, which unwinds mRNA so ribosomes can load on,” Ingolia says. “But why does this drug affect only some mRNAs?” Rocaglamide binds the helicase near this enzyme’s mRNA binding site, so the drug’s proximity to mRNA could affect translation.
Ingolia also wants to know why ribosomes load onto some mRNAs better than others. The answer might be in the hundreds of proteins that bind mRNAs. During transcription, sequence-specific proteins bind and regulate gene expression, and translational regulation may be similar. This has been hard to study until now but “with ribosome profiling, we can ask how proteins affect translation,” he says.
In recognition of his work, Ingolia was selected as a 2014 Rose Hills Innovator, a new UC Berkeley program for up to five early-career faculty with exceptionally high scientific promise. Ingolia also recently received an NIH New Innovator Award.
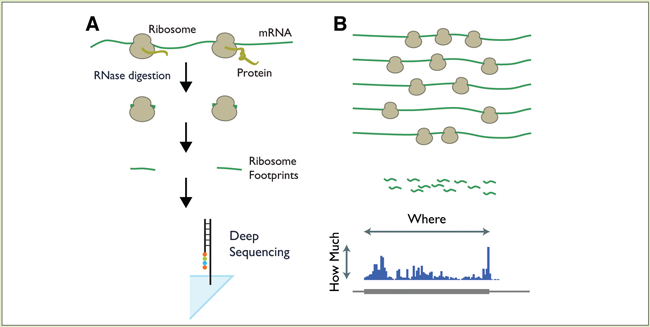
(B) Aligning the footprints to the genome shows which proteins are being synthesized, and at what levels.




