Elçin Ünal

Professor of Genetics, Genomics, Evolution, and Development
Lab Homepage: http://www.unallab.org/Research Interests
My lab aims to define the principles and regulation of meiotic differentiation. This process, also known as gametogenesis, is a conserved developmental program that creates life for the next generation by forming reproductive cells called gametes. Because gametogenesis contains endogenous pathways that prevent age-associated damage from being passed onto progeny, mechanistic dissection of this program offers unique insights into the biology of aging as well as potential therapeutic avenues to combat age-associated diseases including cancer and neurodegeneration. Accordingly, a fundamental question emerges as to how the fitness of gametes is ensured during their production such that they contain the appropriate nuclear and cytoplasmic content to make healthy progeny. Using budding yeast Saccharomyces cerevisiae as a primary model system, we address this question in two complementary frameworks: First, in the context of gene regulation, with the aim of understanding how the essential meiotic processes that ultimately drive cellular rejuvenation are controlled by the meiotic transcriptional program. Second, we study meiotic differentiation in the context of aging, with the aim of understanding how gamete formation promotes cellular rejuvenation and how meiotic cells ensure that age-associated damage such as protein aggregates and dysfunctional organelles are prevented from being transmitted to subsequent progeny. We further extend our studies to multicellular and human cell line systems with an eye on therapeutic potential of our findings.
Current Projects
1. TRANSCRIPTIONAL CONTROL OF REJUVENATION
1a. Leveraging Meiosis-Specific Rejuvenation Pathways to Extend Lifespan
The beneficial effects of gametogenesis on cellular aging and health have been primarily studied in the budding yeast S. cerevisiae. This organism shares striking similarities with metazoans regarding the cellular manifestations of aging including perturbed nuclear and nucleolar integrity, mitochondrial dysfunction, decreased vacuolar/lysosomal acidity and loss of protein homeostasis (Fig. 1). Over time, these types of age-associated damage eventually lead to cell death. Interestingly, when yeast undergoes meiotic differentiation, gametes no longer contain any of the organellar defects originally present in the aged precursor cells. In fact, each of the four gametes is born young, independent of the progenitor’s age (Ünal et al. 2011). These findings demonstrate that meiotic differentiation naturally resets the aging clock, likely by eliminating age-associated damage. 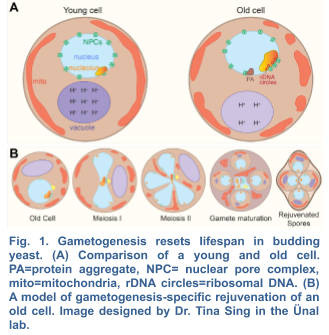
The meiotic transcription factor Ndt80 is the key driver of gamete rejuvenation, however the subset of its gene targets that extend lifespan remains unknown. Based on the finding that ectopic expression of NDT80 increases the longevity of aged somatic cells (Ünal et al. 2011), we have developed a gain-of-function screen to identify additional meiotic genes that extend lifespan when expressed in vegetative cells. To this end, we have generated stage-specific cDNA expression libraries, under the control of an inducible promoter and verified their quality by deep sequencing. In collaboration with Dr. Peter Sudmant at the Department of Integrative Biology, we have developed a rigorous analysis pipeline that utilizes monotonicity z-scores to identify genes that become enriched or disenriched in our screens over time. Our hits from the screen will be further validated by a microfluidic lifespan assay, through our collaboration with Dr. Hao Li's Lab at UCSF. Subsequent functional characterization of these genes in yeast as well as the potential orthologs in C.elegans will pinpoint the cellular pathways and organelles that are limiting for lifespan.
A genome-wide gain-of-function screen for longevity has not been conducted previously. Furthermore, our cDNA libraries include meiosis-specific transcript isoforms that produce novel short ORFs as well as truncated or extended ORF isoforms with potentially altered functions. Such transcripts are not annotated in the standard databases and are therefore not included in any of the commercially available ORF libraries. Therefore, our studies will reveal the function of both known and previously unknown genes in meiotic rejuvenation and healthspan extension. Finally, by performing comparative studies in two separate model systems, we will be able to determine conserved aspects of gametogenesis-induced rejuvenation, which could hold therapeutic potential to counteract cellular aging.
1b. Molecular Mechanism of LUTI-based Repression
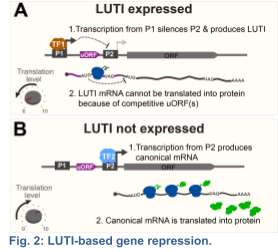
Recent studies suggest that only a fraction of the Ndt80-dependent transcripts are canonical; i.e. fully mapping to one of ~6000 annotated open reading frames (ORFs) within the yeast genome. The rest contain alternative transcription start sites within or outside of the
known ORFs (Brar et al 2012). Among the newly identified meiotic transcripts, my group, in collaboration with Folkert Van Werven's Lab at the Crick Institute, has dissected the mechanism of action for a distinct class that we termed LUTIs – for Long Undecoded Transcript Isoforms (Chen and Tresenrider 2017; Chia et al 2017). LUTIs are expressed from an alternative promoter, which is located upstream of the canonical promoter near the coding region. We found that activation of the LUTI promoter downregulates protein synthesis by a two-pronged mechanism: First, transcription from the LUTI promoter establishes chromatin that silences the canonical promoter in cis, thereby blocking the expression of the protein-coding transcript isoform. Second, the LUTI mRNA cannot be translated into a functional protein, because the upstream open reading frames (uORFs) in its 5’ leader restrict ribosome access to the coding region (Fig. 2). Consequently, a single transcription factor can synchronously activate and repress protein synthesis for distinct sets of genes, depending whether it binds to a canonical or an alternative LUTI promoter, respectively.
To identify new factors involved in LUTI-based regulation, we take an unbiased genetic approach by fusing the 5’ leader of a LUTI-regulated gene in front of a reporter gene, and by selecting mutants that gain the ability to express the reporter despite elevated levels of LUTI expression (LUTI suppressors). By deep sequencing, we then identify such LUTI suppressors and determine their mechanism of action. Our genetic approach is key to uncovering new players in this novel mode of gene regulation.
1c. Evolutionary Conservation of LUTI-based Regulation
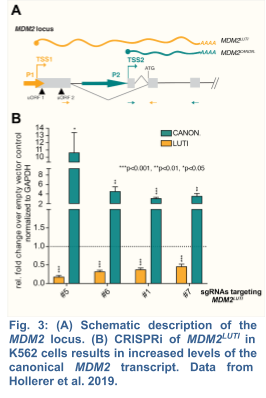
Extending our initial discoveries in yeast, my group, in collaboration with the Brar Lab, has recently demonstrated that the human proto-oncogene MDM2 is regulated by a LUTI (Hollerer et al. 2019) (Fig. 3). As human embryonic stem cells (hESCs) differentiate into endoderm, MDM2LUTI expression is upregulated while the protein-coding MDM2 transcript shows the opposite trend. Because MDM2 is the principal antagonist of p53, a tumor suppressor that promotes mesendodermal gene expression and inhibits pluripotency factors, we posit that MDM2LUTI is important for proper endoderm differentiation. To this end, we plan to test whether the LUTI-based repression requires the same machinery used in yeast. Next, we will determine whether MDM2LUTI is necessary for endoderm differentiation.
Given that at least 40% of the human genes have two or more promoters giving rise to mRNAs with distinct 5’ leaders and translation efficiency (Kimura et al., 2006; Calvo et al., 2009), LUTI-based repression is likely to be a common mode of regulation for many genes besides MDM2. To find those genes, we will expand our studies genome-wide. Initially, we will turn to neural progenitor cell differentiation from hESCs, which has been successfully used for genome-wide RNA and translation measurements (Blair et al., 2017). Once established, we can apply this platform to dissect LUTIs that are induced by Yamanaka transcription factors, which have been recently implicated in human cell rejuvenation (Sarkar et al., 2020).
Using MDM2 regulation as a starting model, these experiments will elucidate the mechanisms involved in LUTI-based repression in human cells. This work will also address the open question of whether LUTI-based regulation serves a functional role in human development. Finally, genome-wide identification of LUTIs will generate new research directions to determine the impact of this gene regulatory mechanism in human development, aging and disease.
2. CELLULAR DRIVERS OF REJUVENATION
2a. Mechanism of Selective Nuclear Inheritance into Gametes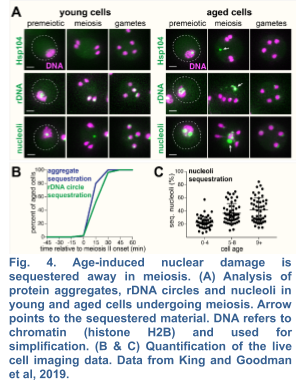
We recently demonstrated the existence of a dramatic nuclear restructuring event in meiosis, which involves the sequestration of nuclear pore complexes (NPCs) into a membranous compartment away from the dividing chromosomes destined for future gametes (Fig. 4 and 5). We named this discarded structure GUNC for Gametogenesis Uninherited Nuclear Compartment (King and Goodman et al. 2019; King and Ünal, 2020). Even though this event is an inherent part of meiosis (i.e. occurring in both young and aged cells), we found it to be essential for the successful elimination of age-associated nuclear defects, thus ensuring gamete rejuvenation. However, the mechanism by which NPCs are sequestered into the GUNC is poorly understood.
Our findings thus far are consistent with a hypothesis that gamete plasma membrane development triggers the formation of a novel scaffold at the rim of growing membranes (Fig. 5), which helps to organize a nuclear envelope diffusion barrier during gametogenesis, thereby culminating in selective nuclear inheritance and subsequent gamete rejuvenation. In order to identify this scaffold, we are undertaking two unbiased approaches: (1) a proximity-dependent biotin labeling approach, which has been successfully applied to determine the constituency of proteins in discrete subcellular compartments, and (2) a genetic screen to identify factors involved in selective nuclear inheritance. Once we identify the candidate factors, we will determine their role in selective nuclear inheritance and gamete rejuvenation.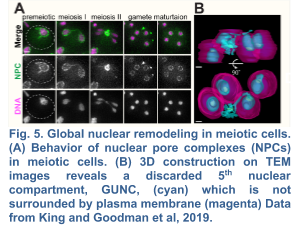
Interestingly, a similar type of NPC remodeling occurs during metazoan spermatogenesis (Ho et al., 2010). Furthermore, several NPC subunits are exceptionally long-lived proteins, accumulating damage over time in yeast, worms and mammals (Sakuma & D’Angelo, 2017). These similarities across such diverse evolutionary lineages suggest an ancestral need to reorganize the nucleus during meiotic differentiation, perhaps for gamete rejuvenation. Thus, our studies could shed light into nuclear quality control pathways in metazoan gametogenesis.
2b. Mitochondrial Inheritance and Quality Control during Gametogenesis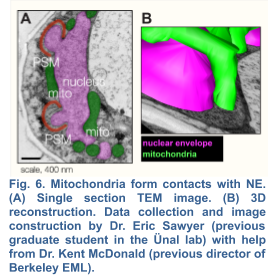
Although a strong link between mitochondrial dysfunction and aging has been established in various model systems, whether and how damaged mitochondria are identified and partitioned in meiosis remains elusive. Evidence suggests that mitochondrial selection occurs in the metazoan germline (Hill et al., 2014; Tworzydlo et al., 2016; Lieber et al., 2019) , however the underlying mechanisms remain largely unexplored. Interestingly in budding yeast, up to 50% of the initial mitochondrial pool is partitioned amongst the four gametes (Brewer and Fangman,1980), while the remaining mitochondria are eliminated by mega autophagy, which involves the programmed lysis of the progenitor cell’s vacuole, an organelle equivalent to the mammalian lysosome (Eastwood et al., 2012). Whether or not there are functional differences between the inherited and discarded mitochondrial populations is currently unknown. Shortly before meiosis II, mitochondria become localized near the nuclei, forming abundant contact sites with the nuclear envelope (Fig. 6). An attractive hypothesis is that the mechanisms that mediate mito-nuclear tethering contribute to limited inheritance of mitochondria during meiosis and therefore could enable selective inheritance of healthier mitochondria. To begin to test this hypothesis, we must first identify the molecular basis of mito-nuclear tethering.
Our data so far suggest that membrane contact sites between mitochondria and outer nuclear envelope/ER are established by novel tethering components that are specific to meiotic cells. To identify the proteins responsible for mito-nuclear tethering, we are undertaking both genetic and biochemical approaches. Following characterization, we will disrupt the mito-nuclear tether(s) and determine how this perturbation affects propagation of healthy mitochondria and gamete rejuvenation.
Both the cellular differentiation program of meiosis and mitochondria are evolutionarily conserved. Perinuclear association of mitochondria occurs in metazoan gametogenesis (Kloc et al., 2004), suggesting further conservation of a mode of meiotic mitochondrial inheritance. Thus, our investigation of mito-nuclear tethering will provide a broader understanding of membrane contact sites and their functions during development, and cellular rejuvenation.
Selected Publications
Research Papers
1. Tresenrider, A., Jorgensen, V., Chia, M., Liao, H., Van Werven, F., Ünal, E. Integrated Genomic Analysis Reveals Key Features of Long Undecoded Transcript Isoform (LUTI)-based Gene Repression. preprint available in BioRxiv 843458
2. Jorgensen, V., Chen, J.1, Vander Wende, H.1, Harris, D., Wong-Deyrup, S.W., Chen, Y., Rangan, P., Brar, G.A., Sawyer, E.M.2, Chan, L.Y.C. 2 and Ünal, E. 2, ¶ (1equal contribution, 2corresponding, ¶ lead contact) Tunable Transcriptional Interference at the Endogenous Alcohol Dehydrogenase Locus in Drosophila Melanogaster. G3 2020 vol. 10 no. 5 1575-1583
3. Chen, J., Liao,A., Powers, E.N., Liao, H., Kohlstaedt, L.A., Evans R., Holly, R.M., Kim Kim, J., Jovanovic, M., Ünal, E. (2020) Aurora B-dependent Ndc80 Degradation Regulates Kinetochore Composition in Meiosis. Genes & Development. 2020 Jan 9 34: 209-225
4. King, G.A.1, Goodman, J.S.1, Schick, J.G., Chetlapalli, K., Jorgens, D., McDonald, K., and Ünal, E. Meiotic Cellular Rejuvenation is Coupled to Nuclear Remodeling in Budding Yeast. eLife 2019 DOI: 10.7554eLife.47156 (1equal contribution)
5. Sawyer, E.M., Joshi, P., Jorgensen, V., Yunus, J., Berchowitz, L.E. and Ünal, E. Developmental Regulation of an Organelle Tether Coordinates Mitochondrial Remodeling in Meiosis. Journal of Cell Biology 2019 DOI: 10.1083/jcb.201807097 (PR)
6. Hollerer, I., Barker, J.C.1, Jorgensen, V.1, Tresenrider, A., Dugast-Darzacq, C., Darzacq, X., Chan, L.Y.C., Tjian, R., Ünal, E. ¶ and Brar, G.A. ¶ Evidence for an Integrated Gene Repression Mechanism based on mRNA Isoform Toggling in Human Cells G3, 2019 vol. 9 no. 4 1045-1053 (1equal contribution, ¶ corresponding)
7. Chen, J., McSwiggen, D.T. and Ünal, E. Single Molecule Fluorescence In Situ Hybridization (Smfish) Analysis in Budding Yeast Vegetative Growth and Meiosis. Journal of Visual Experimentation. 2018 e57774, doi:10.3791/57774
8. Chia, M.H., Tresenrider, A., Chen, J., Spedale, G., Jorgensen, V., Ünal, E.1, and Van Werven F.1. Transcription of a 5’ extended mRNA isoform directs dynamic chromatin changes and interference of a downstream promoter. eLife. 2017 Sep; 14, e27420 (1corresponding)
9. Chen, J.1, Tresenrider, A.1, Chia, M.H., McSwiggen, D.T., Spedale, G., Jorgensen, V., Liao, H., Van Werven, F. 2, and Ünal, E.2 Kinetochore Inactivation by Expression of a Repressive mRNA. eLife. 2017 Sep; 14, e27417 (1equal contribution, 2corresponding)
10. Miller, M.P.1, Ünal, E.1, Brar, G.A., Amon, A. Meiosis I Chromosome Segregation Is Established Through Regulation Of Microtubule-Kinetochore Interactions. eLife. 2012 Dec; 18, e00117 (1equal contribution)11. Ünal, E., Kinde, B., Amon, A. Gametogenesis Eliminates Age-Induced Cellular Damage And Resets Life-Span In Yeast. Science. 2011 Jun; 332, 1554-1557.
Photo credit: Mark Hanson at Mark Joseph Studios.
Last Updated 2020-09-12