Roberto Zoncu

Esther and Wendy Schekman Chair in Basic Cancer Biology, Professor of Molecular Therapeutics
Lab Homepage: http://www.robertozonculab.org/Research Interests
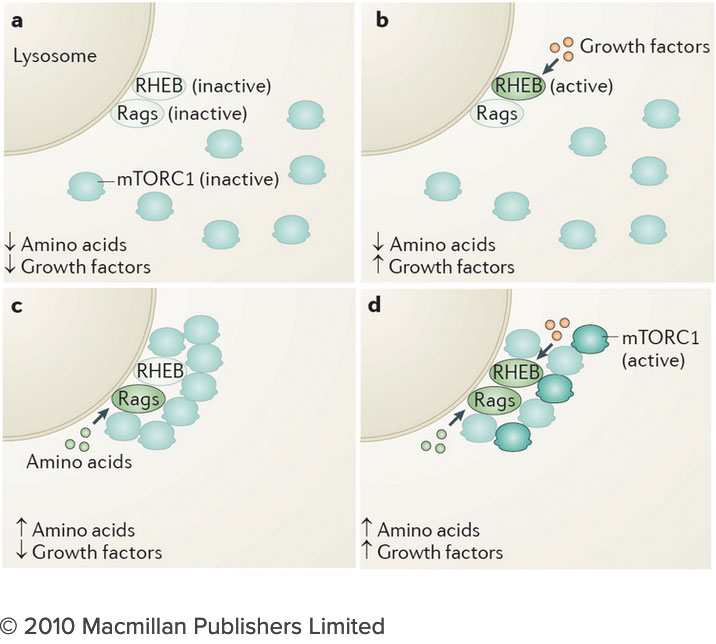 How do the nutrients we consume regulate our growth and homeostasis? Answering this question will help us understand not only how we develop, but also how we age and why we become susceptible to diseases as diverse as cancer, diabetes and neurodegeneration. After decades of research, we know surprisingly little on how nutrients are sensed within cells, and how nutrient-derived signals remodel the cell and enable it to adjust to changing metabolic requirements. Many intracellular compartments, bearing exotic names such as mitochondria, lysosomes and autophagosomes, specialize in storing, releasing and processing metabolites that range from amino acids to sugars, lipids and nucleotides. But how does each organelle sense the quality and quantity of the metabolites it carries? And how is this nutrient and energy information of each organelle communicated to other compartments in the cell?
How do the nutrients we consume regulate our growth and homeostasis? Answering this question will help us understand not only how we develop, but also how we age and why we become susceptible to diseases as diverse as cancer, diabetes and neurodegeneration. After decades of research, we know surprisingly little on how nutrients are sensed within cells, and how nutrient-derived signals remodel the cell and enable it to adjust to changing metabolic requirements. Many intracellular compartments, bearing exotic names such as mitochondria, lysosomes and autophagosomes, specialize in storing, releasing and processing metabolites that range from amino acids to sugars, lipids and nucleotides. But how does each organelle sense the quality and quantity of the metabolites it carries? And how is this nutrient and energy information of each organelle communicated to other compartments in the cell?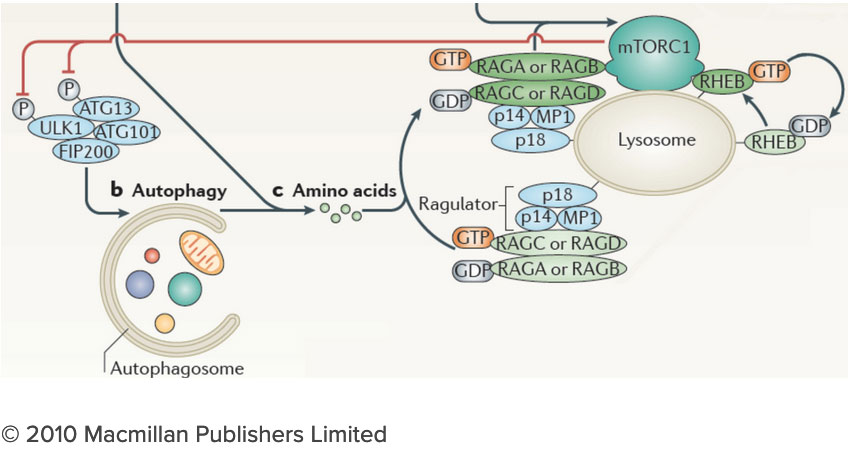 The connection between the lysosome and mTORC1 has important implications for understanding the logic of metabolic regulation. mTORC1 drives biosynthetic processes such as ribosome biogenesis, mRNA translation and lipid synthesis. In turn, the lysosome mediates the breakdown of superfluous or damaged cellular components, thus providing a source of fuel and quality control for the cell. Our findings suggest that localized mTORC1 activation at the lysosome may provide a means for the cell to integrate biosynthetic and catabolic processes in space and time. Thus, elucidating the connection between mTORC1 and the lysosome may point the way to novel approaches for pathologies in which mass accumulation and cellular quality control are deranged, primarily in cancer and protein misfolding diseases.
The connection between the lysosome and mTORC1 has important implications for understanding the logic of metabolic regulation. mTORC1 drives biosynthetic processes such as ribosome biogenesis, mRNA translation and lipid synthesis. In turn, the lysosome mediates the breakdown of superfluous or damaged cellular components, thus providing a source of fuel and quality control for the cell. Our findings suggest that localized mTORC1 activation at the lysosome may provide a means for the cell to integrate biosynthetic and catabolic processes in space and time. Thus, elucidating the connection between mTORC1 and the lysosome may point the way to novel approaches for pathologies in which mass accumulation and cellular quality control are deranged, primarily in cancer and protein misfolding diseases.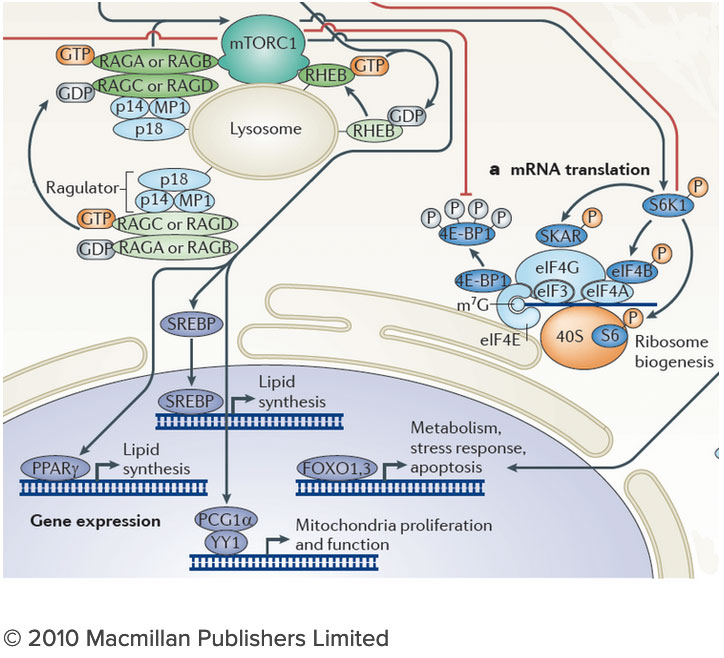 We are investigating the lysosome as a metabolic ‘command and control’ center that i) functions as a signaling hub for nutrient sensing and signaling and ii) controls the storage and delivery of key substrates to the cell’s metabolic pathways. Exploring these exciting directions poses significant challenges. We do not yet have a comprehensive list of the lysosomal resident proteins and internal metabolites. Moreover, we do not understand how the lysosome’s many parts work together to regulate biophysical properties of this organelle such as its internal acidity and membrane potential. We will meet these challenges by developing novel techniques to probe organelle function both in vivo and in vitro, and by integrating them with advanced live cell microscopy and high throughput approaches.
We are investigating the lysosome as a metabolic ‘command and control’ center that i) functions as a signaling hub for nutrient sensing and signaling and ii) controls the storage and delivery of key substrates to the cell’s metabolic pathways. Exploring these exciting directions poses significant challenges. We do not yet have a comprehensive list of the lysosomal resident proteins and internal metabolites. Moreover, we do not understand how the lysosome’s many parts work together to regulate biophysical properties of this organelle such as its internal acidity and membrane potential. We will meet these challenges by developing novel techniques to probe organelle function both in vivo and in vitro, and by integrating them with advanced live cell microscopy and high throughput approaches.Current Projects
MECHANISMS OF NUTRIENT SIGNAL INTEGRATION AT THE LYSOSOME
The molecular mechanisms through which cells sense nutrients remain largely unknown, but their elucidation is key to our understanding of metabolic regulation both in normal and disease states. At the center of nutrient sensing and growth regulation is an ancient protein kinase known as the mechanistic Target of Rapamycin Complex 1 (mTORC1). In response to nutrient abundance, mTORC1 physically translocates from the cytosol to the lysosomal membrane, where its kinase function becomes activated. A central aspect of mTORC1 function that has so far remained poorly understood is its ability to integrate chemically diverse nutrient inputs, and to translate these combined signals into optimal balance of anabolic and catabolic programs.
We recently discovered that mTORC1 specifically senses a cellular lipid, cholesterol, at the lysosome. Cholesterol is a key structural component for cellular membranes and precursor for important hormones, but whether and how cholesterol can cross-talk with pathways that regulate cellular metabolism was unknown. We found that cholesterol triggers the recruitment of mTORC1 from the cytoplasm to the surface of lysosomes, concomitant with stimulation of the kinase activity of mTORC1. This effect of cholesterol requires the Rag GTPases, which serve as a bridge between mTORC1 and the lysosomal surface.
Combining targeted manipulations of the lipid content of selected organelle populations with functional assays for mTORC1 activation, we are elucidating key aspects of this newly identified signaling pathway. In particular, we are investigating i) the identity of cholesterol-binding proteins that activate or inhibit mTORC1 ii) the molecular mechanisms that translate cholesterol levels into Rag GTPase regulation and iii) the physiological and pathogenic roles of the cholesterol-mTORC1 axis. In particular, our work is illuminating how aberrant cholesterol sensing by mTORC1 could disrupt cellular quality control and drive the metabolic and neurodegenerative disease, Niemann-Pick type C.
INTER-ORGANELLE COMMUNICATION IN METABOLIC CONTROL
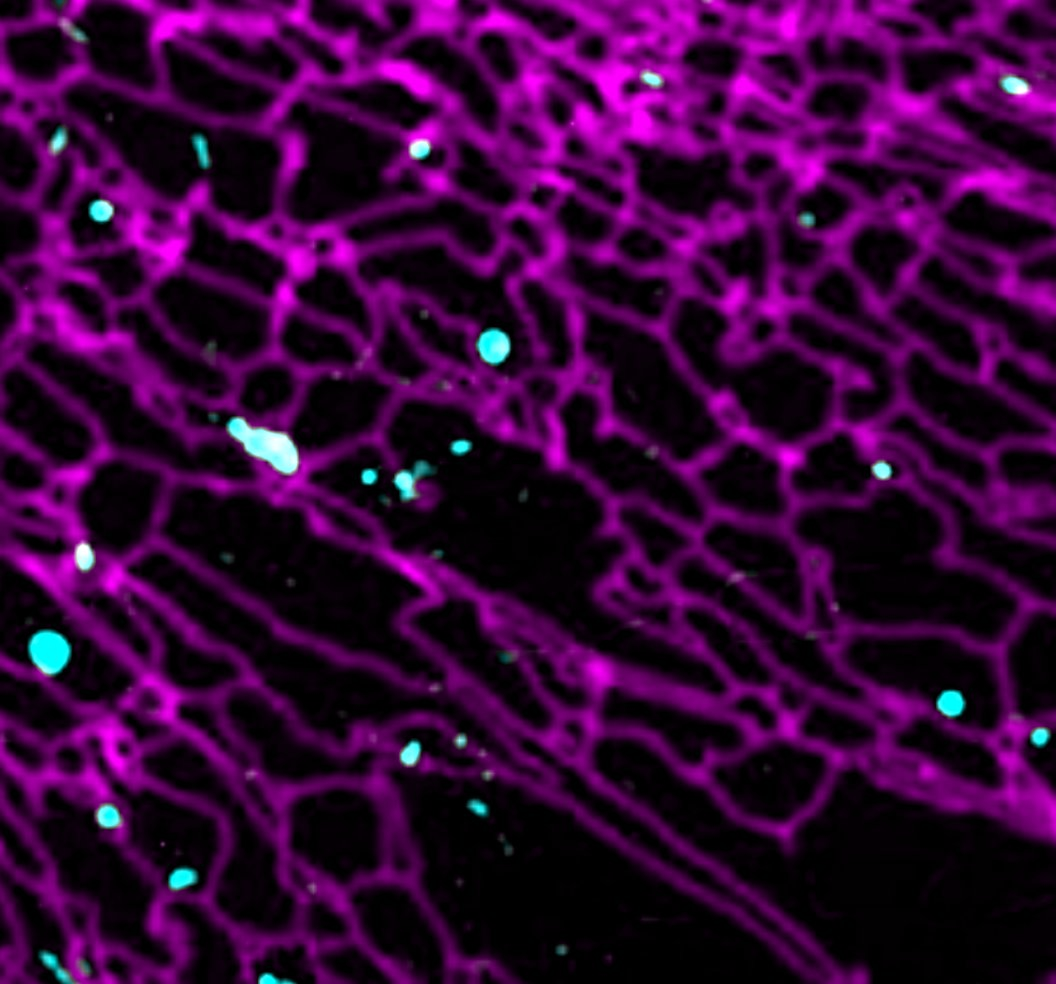
In order to execute its pro-anabolic and pro-catabolic programs, the lysosome engages in extensive interactions, both physically and functionally, with other membrane-bound organelles. For example during autophagy, a 'self-eat' process that clears superfluous or damaged cellular components, lysosomes fuse with double-membraned autophagosomes, allowing the cargo molecules captured by the autophagosome to be delivered to the lysosomal lumen for degradation. However, the factors that regulate autophagosome-lysosome interaction remain poorly understood.
In addition to direct fusion, the lysosome engages in stable physical junctions known as membrane contact sites (MCSs) with other membrane compartments. The MCSs provide the lysosome with a direct route for the exchange lipids, ions and other metabolically relevant signals. However, the full range of inter-organelle interactions, and how they impact lysosome-dependent metabolic programs (i.e. mTORC1 signaling) are wide-open questions. Combining live cell microscopy, high-throughput genetic screens and mass spectrometry-based profiling of specific organelle populations, we are conducting in-depth investigations of how the lysosome interfaces with other important organelles such as ER, autophagosomes, mitochondria and lipid droplets. Our ultimate goal is to discover organelle-based metabolic circuits and dissect their physiological roles in different cellular and physiological contexts. An especially exciting avenue is understanding how cancer cells rewire the composition and function of their organellar circuitries in order to achieve growth and adaptive advantages.
BOTTOM-UP RECONSTITUTION AND CHEMICAL MANIPULATION OF LYSOSOMAL SIGNALING
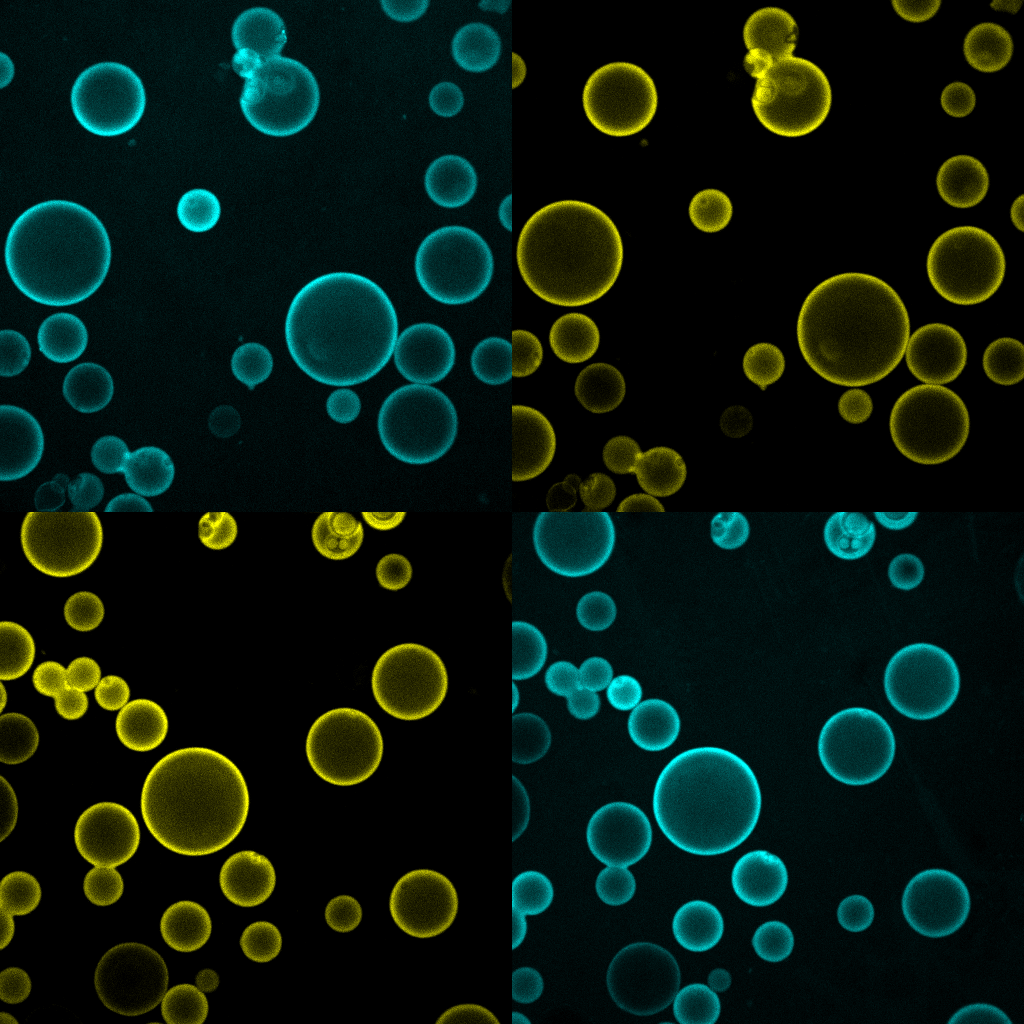
As cell biologists and biochemists, we believe that one cannot truly understand a biological process unless some, or all, of its key features can be recreated in a test tube. This is especially true for lysosomal nutrient signaling, a process of remarkable complexity that involve the precise orchestration of multiple protein complexes, membrane trafficking steps, and metabolite transport rates, all in response to rapidly changing metabolic conditions. In order to progress toward the creation of a 'synthetic lysosome', we combine biochemical reconstitution of proteins and vesicles within in vitro systems that we characterize using advanced live microscopy (bulk and single molecule). These systems are informed by structural insights (derived from X-ray crystallography and cryo-EM) that we developed through collaborative work, thus they take into account stoichiometric and topological constraints likely to occur in an intact cell (article 1 and article 2).
A deep mechanistic understanding of the lysosomal nutrient-sensing system can also inform the design of innovative chemical screening platforms that can enable precise manipulation of nutrient signaling. This is especially critical in the context of mTORC1, given that currently available inhibitors are incomplete (rapamycin) or likely to cause significant toxicities (ATP-competitive). In collaborative work, we have employed covalent chemistry to identify novel mTORC1 inhibitors that operate by targeting the protein complexes that recruit mTORC1 to the lysosomal membrane. Ultimately, our reconstitution and chemical screening pipeline may lead to the identification of novel compounds to combat cancer, protein misfolding disease and metabolic syndrome.
Selected Publications
Lawrence, R.E., Fromm, S.A., Fu, Y., Yokom, A.L., Kim, D.J., Thelen, A.M., Young, L.N., Lim, C.-Y., Samelson, A.J., Hurley, J.H*, Zoncu, R*, 2019. Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex. Science. https://doi.org/10.1126/science.aax0364
Lim, C.-Y., Davis, O.B., Shin, H.R., Zhang, J., Berdan, C.A., Jiang, X., Counihan, J.L., Ory, D.S., Nomura, D.K., Zoncu, R., 2019. ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat. Cell Biol. 21, 1206–1218. https://doi.org/10.1038/s41556-019-0391-5.
Chung CY, Shin HR, Berdan CA, Ford B, Ward CC, Olzmann JA, Zoncu R*, Nomura DK*. Covalent targeting of the vacuolar H+-ATPase activates autophagy via mTORC1 inhibition. Nat Chem Biol. 2019 Aug;15(8):776-785.
Lawrence, R.E, Zoncu, R. The lysosome as a cellular center for signaling, metabolism and quality control. Nat Cell Biol. 2019 Feb;21(2):133-142.
Lawrence RE, Cho KF, Rappold R, Thrun A, Tofaute M, Kim DJ, Moldavski O, Hurley JH, Zoncu R. A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase-Ragulator lysosomal scaffold. Nat Cell Biol. 2018;20(9):1052-63.
Su MY, Morris KL, Kim DJ, Fu Y, Lawrence R, Stjepanovic G, Zoncu R*, Hurley JH*. Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex. Mol Cell. 2017;68(5):835-46 e3. PMCID: PMC5722659.
Thelen AM, Zoncu R. Emerging Roles for the Lysosome in Lipid Metabolism. Trends Cell Biol. 2017
Nguyen TB, Louie SM, Daniele JR, Tran Q, Dillin A, Zoncu R, Nomura DK, Olzmann JA. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Developmental cell. 2017;42(1):9-21 e5.
Di Malta C, Siciliano D, Calcagni A, Monfregola J, Punzi S, Pastore N, Eastes AN, Davis O, De Cegli R, Zampelli A, Di Giovannantonio LG, Nusco E, Platt N, Guida A, Ogmundsdottir MH, Lanfrancone L, Perera RM, Zoncu R, Pelicci PG, Settembre C, Ballabio A. Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science. 2017;356(6343):1188-92.
Castellano BM, Thelen AM, Moldavski O, Feltes M, van der Welle RE, Mydock-McGrane L, Jiang X, van Eijkeren RJ, Davis OB, Louie SM, Perera RM, Covey DF, Nomura DK, Ory DS, Zoncu R. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 2017;355(6331):1306-11.
Belyy, V., Shih, S.M., Bandaria, J., Huang, Y., Lawrence, R.E., Zoncu, R., and Yildiz, A. (2017). PhotoGate microscopy to track single molecules in crowded environments. Nat Commun 8, 13978.
Perera RM*, and Zoncu R*. (2016). The Lysosome as a Regulatory Hub. Annu Rev Cell Dev Biol.
Young, L.N., Cho, K., Lawrence, R., Zoncu, R., and Hurley, J.H. (2016). Dynamics and architecture of the NRBF2-containing phosphatidylinositol 3-kinase complex I of autophagy. Proceedings of the National Academy of Sciences of the United States of America 113, 8224-8229.
Last Updated 2019-11-15