Rebecca Heald

Flora Lamson Hewlett Chair, Professor of Cell Biology, Development and Physiology*
*and
Research Interests
My laboratory’s ongoing research explores two fundamental areas of biology: cell division and size control. Our unique and interdisciplinary approaches take advantage of in vitro systems, particularly cytoplasmic extracts prepared from eggs of the frog Xenopus laevis that reconstitute mitotic chromosome condensation and spindle assembly and function in vitro. To study mechanisms of spindle and organelle size control, we have utilized a smaller, related frog, Xenopus tropicalis, to investigate interspecies scaling, and extracts prepared from fertilized eggs at different stages of embryogenesis to study developmental scaling. Our research has provided novel insight into cell division and morphogenesis, processes essential for viability and development, and defective in human diseases such as cancer. Current research is focused on two major areas. The first is to elucidate novel mitotic mechanisms that promote accurate chromosome segregation, characterize the key players that define spindle architecture, and advance reconstitution experiments toward a systems-level understanding of the spindle. The second is to investigate size control mechanisms at the subcellular, cellular and organism levels, developing tools for chromosome labeling and mutagenesis in Xenopus, and leveraging phylogenetic relationships among different frog species. Our research is highly collaborative and will provide new insight into the underlying principles of spindle assembly and biological size control, as well as the molecular basis of variation that contributes to genomic instability and evolution.
Current Projects
Cell Division. Mitosis is arguably the most dramatic event in the life of a cell. Chromosomes condense, organelles vesiculate, and the microtubule cytoskeleton rearranges into a bipolar spindle that attaches to chromosomes and segregates a complete set to each daughter cell. Although the morphological changes that occur during mitosis were first observed over a century ago, we still do not understand how these dynamic events are orchestrated. We develop assays in Xenopus egg extracts to elucidate mitotic events and identify novel players. The extract system is ideal because it provides the complexity of total cytoplasm, yet is open, allowing control of cell cycle progression, specific inactivation of individual components, high-resolution observation, and biochemical and biophysical approaches. We take advantage of this system to address fundamental questions: 1. How do chromosomes condense during mitosis? 2. How do biochemical signals from chromosomes contribute to spindle assembly? 3. How does the spindle assemble and adapt to different cellular contexts, and 4. What is the contribution of cytoplasmic membranes and how are they reorganized and distributed? These projects inform our ultimate objective: 5. Can we reconstitute the spindle from purified components?
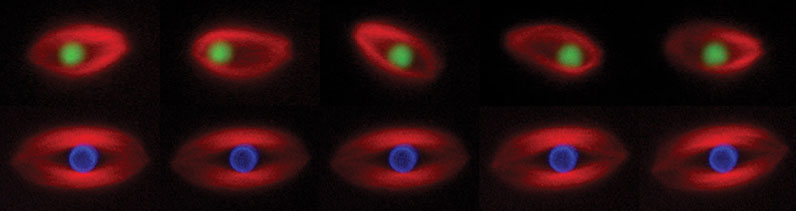
Figure 1. Time-lapse images of spindles assembled in Xenopus egg extract around a bead coated with RCC1, the guanine nucleotide exchange factor for Ran (green, upper panel) or a bead coated with chromatin (blue, lower panel). This experiment shows that a single protein (RCC1) can induce spindle assembly, but the bead oscillates within the spindle.
Size Control. Size varies widely in biology at many levels: the animal, the cells that make up the animal, and the contents of the cells, but we don’t understand how scaling occurs so that everything fits and functions properly. Correct organelle size is crucial for cell function, architecture, and division, but the control systems that a cell uses to regulate the size of its organelles are virtually unknown. Macromolecular cellular assemblies like the ribosome or nuclear pore self assemble from a fixed number of subunits and do not vary in size. In contrast, many complex organelles adjust in size to suit a particular cellular environment. Loss of certain tumor suppressors, or overexpression of oncogenes such as myc can result in an increase in cell and nuclear size. Although size-related cell morphology changes are characteristic of cancer pathology and used diagnostically, the precise nature of these changes and their underlying molecular mechanisms are poorly understood.
Xenopus frogs offer two physiological contexts in which we can investigate organelle size control. First, we can compare Xenopus laevis to the smaller, related species Xenopus tropicalis, which lays smaller eggs and has proportionally smaller cells throughout development. Second, we can compare different stages of Xenopus laevis embryogenesis, as the ~1 millimeter diameter egg rapidly cleaves to form smaller blastomeres. A unique aspect of our approach has been to prepare cytoplasmic extracts from eggs and embryos that recapitulate organelle scaling in vitro, which we can use to identify molecular differences that underlie size changes. Together with the laboratory of Dan Fletcher, we have engineered synthetic cells of defined sizes and lipid composition by encapsulating Xenopus cytoplasm inside droplets.
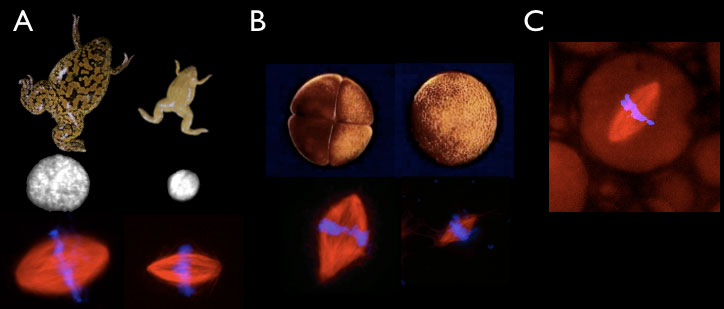
Figure 2. In vitro scaling systems. A. Xenopus laevis (left) and Xenopus tropicalis (right) egg extracts generate different sized nuclei and mitotic spindles in vitro. (B) Extracts can be prepared from embryos at the 4 cell (left) or 4000 cell (right) embryos that form different-sized spindles in vitro. (C) Spindle assembly reactions can be performed inside different-sized droplets of cytoplasm in oil.
Selected Publications
Brownlee C and Heald R. (2019) Importin alpha partitioning to the plasma membrane regulates intracellular scaling. Cell doi: 10.1016/j.cell.2018.12.001. PMID30639102
Gibeaux R, Acker R, Kitaoka M, Georgiou G, van KruijsbergenI, Ford B, Marcotte EM, Nomura DK, Kwon T, Veenstra GJC, Heald R. (2018) Paternal chromosome loss and metabolic crisis contribute to hybrid inviability in Xenopus. Nature 553, 337-341. PMC5988642
Grenfell AW, Heald R, Strzelecka M. (2016) Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J Cell Biol 214, 133-41. PMC4949451
Lane AB, Strzelecka M, Ettinger A, Grenfell AW, Wittmann T, Heald R. (2015) Enzymatically generated CRISPR libraries for genome labeling and screening. Dev Cell 34, 373-8. PMC4536113
Crowder ME, Strzelecka M, Wilbur JD, Good MC, von Dassow G, Heald R. (2015) A comparative analysis of spindle morphometrics across metazoans. Curr Biol 25, 1542-50. PMC4464779
Helmke KJ, Heald R. (2014) TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J Cell Biol 206, 385-93. PMC4121975
Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. (2013) Cytoplasmic volume modulates spindle size during embryogenesis. Science 342, 856-60. PMC4094345
Wilbur J, Heald R. (2013) Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife 2013;2:e00290.
Schlaitz A-L, Thompson J, Wong CCL, Yates JR, Heald R. (2013) REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell 26, 316-323. PMC3745822
Halpin D, Kalab P, Wang J, Weis K, Heald R. (2011) Spindle assembly around RCC1 coated beads in Xenopus egg extracts. PLoS Biol 9, e1001225. PMC3246454
Loughlin R, Wilbur JD, McNally FJ, Nedelec F, Heald R. (2011) Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147, 1937- 1407. PMC3240848
Photo credit: Mark Hanson at Mark Joseph Studios.
Last Updated 2019-01-18