Iswar Hariharan

Associate Dean of Academic Affairs, William V. Power Chair in Biology and American Cancer Society Research Professor, Professor of Cell Biology, Development and Physiology
Lab Homepage: http://mcb.berkeley.edu/labs/hariharan/Research Interests
Our laboratory studies the mechanisms that regulate growth at the level of individual cells as well as the entire organism. We are interested in the growth that occurs during development to determine the eventual size and form of an organism. We are also interested in the mechanisms by which damaged tissue is replaced as a result of regenerative growth.
To understand the way that growth is regulated in both of these situations, we conduct genetic studies in the fruitfly, Drosophila melanogaster, to identify genes that regulate growth, cell proliferation and cell death. In addition to identifying some of the key regulators of tissue growth in Drosophila, our studies have also helped identify genes that are mutated in human cancers.
Current Projects
1) Regulation of growth in imaginal discs during development
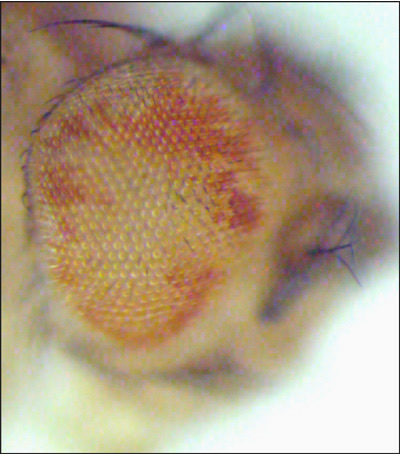
Over the last ten years, we have conducted a genetic screen that has led to the identification of many genes that restrict tissue growth and cell proliferation in vivo. Following mutagenesis, we generated flies with eyes that contain both wild-type tissue (marked red) and mutant tissue (marked white). By comparing the relative representation of the two types of tissue we can identify mutations that allow mutant cells to outgrow their wild type neighbors.
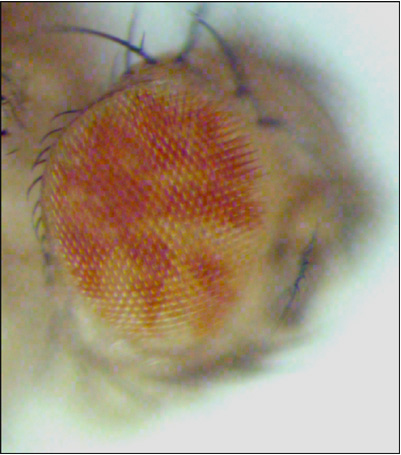 |
The mutant eye (right image) shows an increase in the relative representation of mutant tissue (white) when compared to the control (left image). |
 |
Once we identify the mutant flies, we map the mutation and characterize the mutated gene at the molecular level. Further studies of the encoded protein usually provide clues as to how this gene functions to regulate tissue growth. Our eventual aim is to understand the cellular circuitry that regulates the growth process.
Genes identified in our screen include the orthologs of the genes mutated in the human disease tuberous sclerosis (Tsc1 and Tsc2), pten, components of the RTK/Ras pathway (e.g. Gap1, argos, capicua ) as well as many genes that function to regulate growth in ways that are still not well understood. One of the genes that we have characterized archipelago, encodes a protein that facilitates the destruction of Cyclin E and Myc. Another set of three genes (salvador, warts and hippo) constitute a novel pathway that restricts tissue growth. For the human orthologs of archipelago and salvador, we identified mutations in cancer cell lines. We and others have identified mutations in the human ortholg of archipelago (hCdc4, FBW7) in primary human tumors.
2) Studies of regeneration in Drosophila
The fundamental question in the study of regeneration is why some animals have a remarkable capacity to regenerate damaged tissues while others do not. For instance, while urodele amphibians (salamanders, newts) are capable of regenerating their limbs after amputation, mammalian limbs have very little regenerative capacity. Similarly, in many species, less regenerative capacity is observed in adults than at earlier stages of development. The long term goal of our research is to understand the genetic pathways that regulate the capacity of a tissue for regenerative growth.
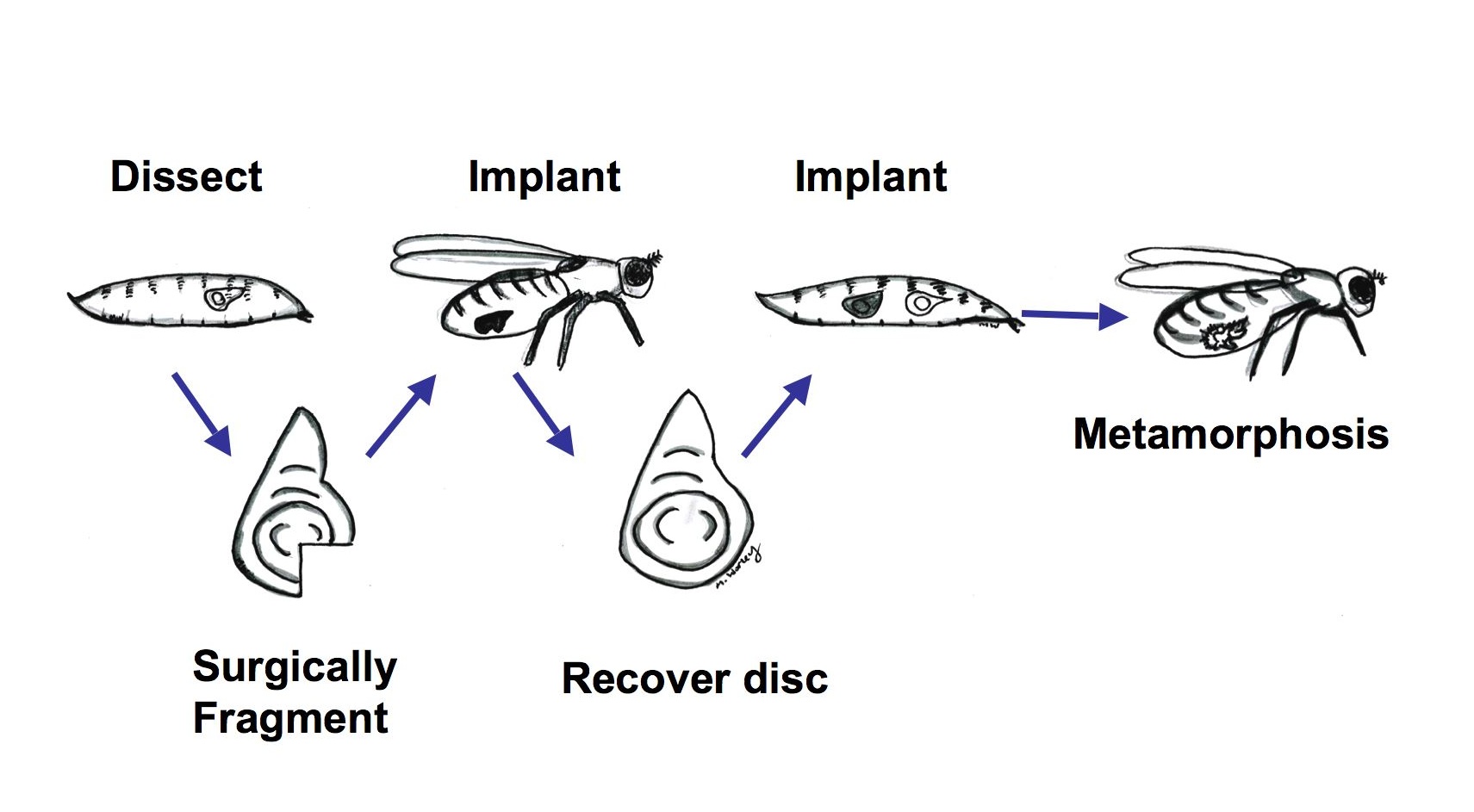
The imaginal discs of Drosophila larvae are capable of regenerating missing portions when implanted into the abdomen of adult females. However, these experiments are technically difficult and not easily incorporated into large-scale genetic screens.
 |
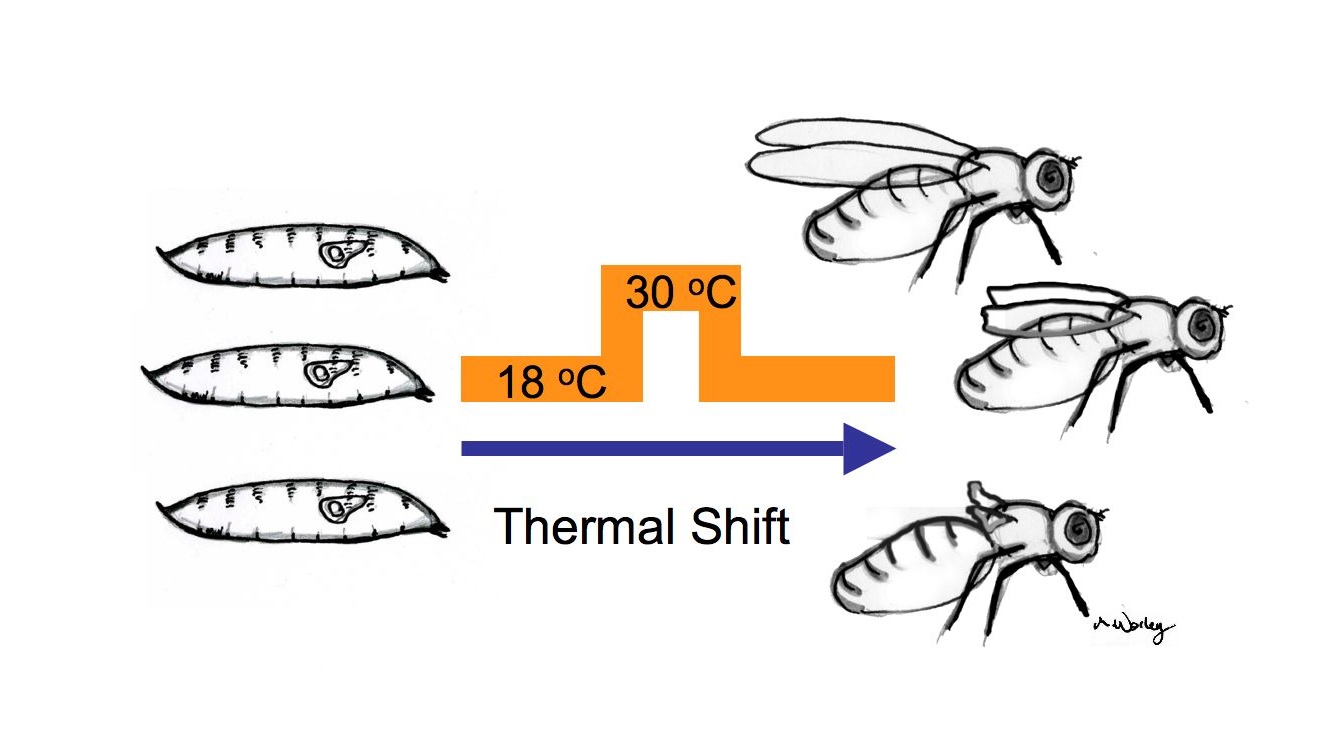 |
| The traditional way of studying imaginal disc regeneration | Our system allows the study of imaginal disc regeneration in situ |
(Drawings by Melanie Worley)
To take advantage of the powerful tools of fly genetics, we have devised a way of studying tissue regeneration in Drosophila that does not require surgical manipulations. Using a temperature shift, we are able to ablate the portion of the larval wing imaginal disc that gives rise to the wing blade of the adult, and to study the ensuing regenerative growth. We can easily score the extent of regeneration by examining the size of the wings of adult flies. Our laboratory is currently using this method to study the properties of regenerative growth and to conduct genetic screens for genes that regulate tissue regeneration.
We have also addressed the mechanism by which tissue damage in larvae delays pupariation. Our studies indicate that tissue damage activates a developmental checkpoint that delays the production of the peptide hormone PTTH. Since PTTH activates the release of ecdysone by the ring gland and promotes metamorphosis, delaying PTTH production extends the larval stage - a condition that is permissive for regenerative growth. Our results point to a role for retinoids in the mechanism that links tissue damage to delayed PTTH production. This masy provide a tractable model for understanding how individual tissues communicate the presence of damage to the entire organism.
Selected Publications
Cruz J, Sun WY, Verbeke A, Hariharan IK. Single-cell transcriptomics of X-ray irradiated Drosophila wing discs reveals heterogeneity related to cell-cycle status and cell location. bioRxiv [Preprint]. 2025 Feb 11:2024.12.10.627868. doi: 10.1101/2024.12.10.627868. PMID: 39990483; PMCID: PMC11844406.
Spitzer DC, Sun WY, Rodríguez-Vargas A, Hariharan IK. The cell adhesion molecule Echinoid promotes tissue survival and separately restricts tissue overgrowth in Drosophila imaginal discs. bioRxiv [Preprint]. 2023 Aug 6:2023.08.04.552072. doi: 10.1101/2023.08.04.552072. PMID: 37577631; PMCID: PMC10418178.
Friesen S, Hariharan IK. Coordinated growth of linked epithelia is mediated by the Hippo pathway. bioRxiv [Preprint]. 2023 Mar 17:2023.02.26.530099. doi: 10.1101/2023.02.26.530099. PMID: 36993542; PMCID: PMC10054945.
Worley MI, Everetts NJ, Yasutomi R, Chang RJ, Saretha S, Yosef N, Hariharan IK. Ets21C sustains a pro-regenerative transcriptional program in blastema cells of Drosophila imaginal discs. Curr Biol. 2022 Aug 8;32(15):3350-3364.e6. doi: 10.1016/j.cub.2022.06.040. Epub 2022 Jul 11. PMID: 35820420; PMCID: PMC9387119.
Everetts NJ, Worley MI, Yasutomi R, Yosef N, Hariharan IK. Single-cell transcriptomics of the Drosophila wing disc reveals instructive epithelium-to-myoblast interactions. Elife. 2021 Mar 22;10:e61276. doi: 10.7554/eLife.61276. PMID: 33749594; PMCID: PMC8021398.
Harris RE, Stinchfield MJ, Nystrom SL, McKay DJ, Hariharan IK. Damage-responsive, maturity-silenced enhancers regulate multiple genes that direct regeneration in Drosophila. Elife. 2020 Jun 3;9:e58305. doi: 10.7554/eLife.58305. PMID: 32490812; PMCID: PMC7299344.
Harris RE, Setiawan L, Saul J, Hariharan IK. Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. Elife. 2016 Feb 3;5:e11588. doi: 10.7554/eLife.11588. PMID: 26840050; PMCID: PMC4786413.
Hariharan IK. Organ Size Control: Lessons from Drosophila. Dev Cell. 2015 Aug 10;34(3):255-65. doi: 10.1016/j.devcel.2015.07.012. PMID: 26267393; PMCID: PMC4547687.
Hariharan IK, Wake DB, Wake MH. Indeterminate Growth: Could It Represent the Ancestral Condition? Cold Spring Harb Perspect Biol. 2015 Jul 27;8(2):a019174. doi: 10.1101/cshperspect.a019174. PMID: 26216720; PMCID: PMC4743077.
Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev Cell. 2009 Jun;16(6):797-809. doi: 10.1016/j.devcel.2009.04.015. PMID: 19531351; PMCID: PMC2705171.
Tseng AS, Tapon N, Kanda H, Cigizoglu S, Edelmann L, Pellock B, White K, Hariharan IK. Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signaling pathway. Curr Biol. 2007 Apr 17;17(8):728-33. doi: 10.1016/j.cub.2007.03.023. Epub 2007 Mar 29. PMID: 17398096; PMCID: PMC2699483.
Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003 Aug 22;114(4):457-67. doi: 10.1016/s0092-8674(03)00557-9. PMID: 12941274.
Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002 Aug 23;110(4):467-78. doi: 10.1016/s0092-8674(02)00824-3. PMID: 12202036.
Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001 Sep 20;413(6853):311-6. doi: 10.1038/35095068. PMID: 11565033.
Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001 May 4;105(3):345-55. doi: 10.1016/s0092-8674(01)00332-4. PMID: 11348591.
Last Updated 2025-06-10