Eva Nogales

Howard Hughes Medical Institute Investigator,
Professor of Biochemistry, Biophysics and Structural Biology*
*And Affiliate, Division of Cell and Developmental Biology And Chair, Graduate Group in Biophysics
Research Interests
Our lab is dedicated to gaining mechanistic insight into crucial molecular processes in the life of the eukaryotic cell. Our two main research topics are the role of cytoskeletal dynamics in cell division, and the molecular mechanisms governing the flow of genetic information. The unifying principle in our work is the emphasis on studying macromolecular assemblies as whole units of molecular function via visualization of their functional states and regulatory interactions. We use electron microscopy (EM) and image analysis, biochemical and biophysical assays, towards a molecular understanding of our systems of interest.
Current Projects
1 –Structural Basis and Regulation of Microtubule Dynamic Instability
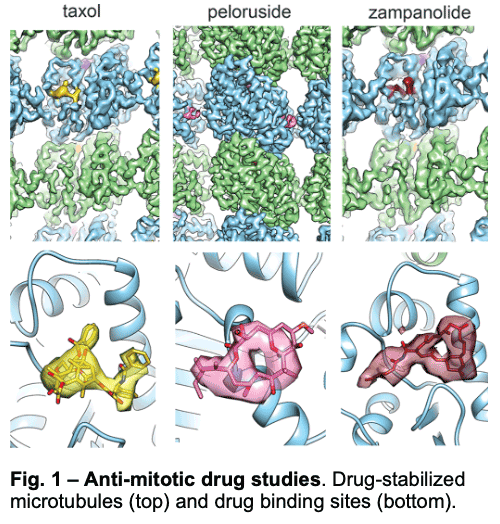 1.1 – Drug Binding and Effects
1.1 – Drug Binding and Effects
Microtubules are essential cytoskeletal polymers involved in a myriad of cellular functions that are regulated by numerous associated proteins and constitute a major target for cancer treatment. Our lab continues to break new ground on the characterization of their structure and regulatory interactions.
We have visualized drugs that stop mitosis through the stabilization of microtubules. We showed that while peloruside acts at lateral contacts and regularizes the microtubule lattice, Taxol and zampanolide affect the longitudinal interface between tubulin dimers and induce microtubule heterogeneity (Fig. 1). These findings suggested the use of combinations of microtubule-stabilizing agents to regulate microtubule activity and improve therapeutic potential.
1.2 – Regulation by Nucleotide and Associated Proteins
We are probing the distinct effects of nucleotide and microtubule-associated factors on the structure of the microtubule. For example:
We have described the interactions between the calponin homology domain of the +tip-tracking protein EB3 and the four tubulin subunits it contacts (Fig. 2, orange), and proposed that EB (End-Binding) proteins modulate structural transitions at the growing microtubule end by recognizing and promoting an intermediate state in the GTP hydrolysis cycle.
-
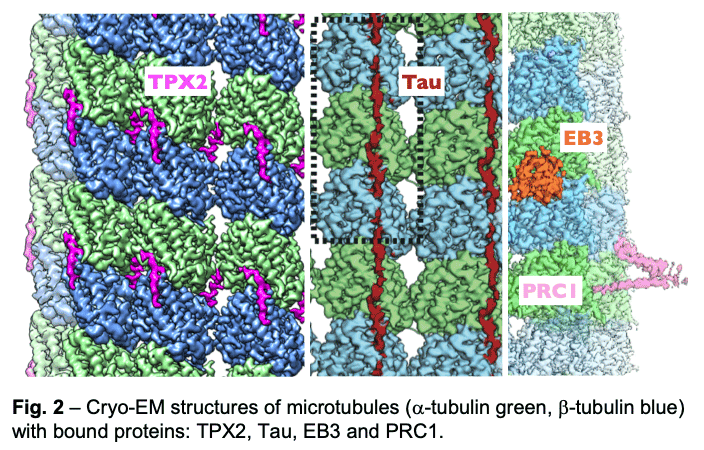 Our study of TPX2 (Fig. 2, magenta) explains both the recognition of the GTP-form of tubulin and the stabilizing effect of TPX2. TPX2 uses two flexibly linked elements to bind simultaneously across longitudinal and lateral tubulin interfaces. These elements overlap with the binding site of importins on TPX2, explaining how the Ran-GTP gradient can regulate TPX2-dependent MT formation.
Our study of TPX2 (Fig. 2, magenta) explains both the recognition of the GTP-form of tubulin and the stabilizing effect of TPX2. TPX2 uses two flexibly linked elements to bind simultaneously across longitudinal and lateral tubulin interfaces. These elements overlap with the binding site of importins on TPX2, explaining how the Ran-GTP gradient can regulate TPX2-dependent MT formation. - Of special notice is our study of the neuronal Tau protein. Alzheimer's progression is associated with the accumulation of neurofibrillary tangles of tau that follows phosphorylation of this protein and its dissociation from microtubules. Given the involvement of tau in this and other neurological diseases (taupathies), both the amyloid and physiological structure of tau bound to the microtubules had been pursued for years. Our studies showed how tau staples tubulin subunits on the surface of microtubules, thus explaining its effect stabilizing the polymer (Fig. 2, maroon). The most tightly bound segment identified in our structure includes a clinically relevant site of tau phosphorylation, explaining the competition between microtubule interaction and tau aggregation. We continue to study regulatory microtubule binders with an emphasis on combinatorial interactions that read or determine the structural states at microtubule ends at the heart of dynamic instability and microtubule function.
2 –Regulation of gene expression
Transcriptional regulation of gene expression is critical for growth and survival. We study epigenetic gene silencing by PRC2 (Polycomb Repressive Complex 2) and transcription initiation by the Pol II system. The molecular machinery involved in the latter include a number of challenging multi-subunit complexes that are conformationally and biochemically complex, and only accessible in minute amounts due to their scarcity in the cell.
2.1 – PRC2 and Gene Silencing
- PRC2 establishes transcriptional repression by tri-methylating Lysine 27 of histone H3, a process mediated by cofactors such as AEBP2 or JARID2. We have now obtained the high-resolution structure of a full complex and showed how AEBP2 stabilizes PRC2 by stapling the top and bottom lobes, and how both cofactors mimic histone tails in their binding to the WD40 domains within the core PRC2.
-
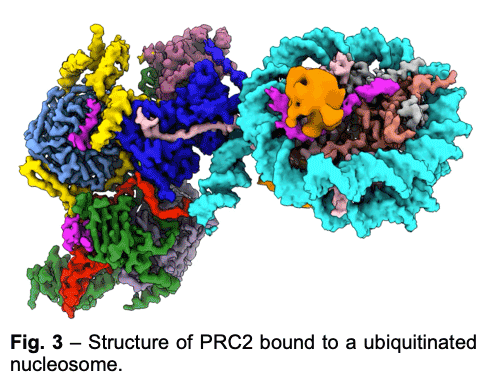 PRC2 is stimulated by its own catalytic product and is more active on dinucleosomes than H3 tails or single nucleosomes. We visualize PRC2 bound to a dinucleosome that would correspond to a boundary condition: one nucleosome being unmodified and the other methylated. We showed how a single PRC2, via interactions with nucleosomal DNA, positions the H3 tails of the activating and substrate nucleosome to interact with EED and the SET domain of EZH2, respectively. We also showed that the geometry of PRC2-DNA interactions allows PRC2 to accommodate varying lengths of linker DNA between nucleosomes.
PRC2 is stimulated by its own catalytic product and is more active on dinucleosomes than H3 tails or single nucleosomes. We visualize PRC2 bound to a dinucleosome that would correspond to a boundary condition: one nucleosome being unmodified and the other methylated. We showed how a single PRC2, via interactions with nucleosomal DNA, positions the H3 tails of the activating and substrate nucleosome to interact with EED and the SET domain of EZH2, respectively. We also showed that the geometry of PRC2-DNA interactions allows PRC2 to accommodate varying lengths of linker DNA between nucleosomes. - One proposed model of PRC2 recruitment to genomic loci is via the recognition of PRC1-mediated mono-ubiquitination of histone H2A at lysine 119 (H2AK119ub1). Our cryo-EM structure of human PRC2 with cofactors JARID2 and AEBP2 bound to a nucleosome substrate containing H2AK119ub1 shows that JARID2 and AEBP2 each interact with one of the two ubiquitin molecules in the nucleosome. Biochemical assays demonstrate a secondary activation of PRC2 by JARID2 and AEBP2 on H2AK119ub1-containing nucleosomes besides the primary EED-mediated allosteric activation of PRC2 by methylated JARID2. Furthermore, we also found that the joint presence of JARID2 and AEBP2 partially reduces the inhibition of PRC2 methyltransferase activity by the transcriptionally active histone post-translational modifications H3K4me3 and H3K36me3. Thus, our work also suggests that JARID2 and AEBP2 are likely to play a key role in regulating PRC2 activity on genomic regions with active transcription marks.
2.2 – Transcription Initiation
-
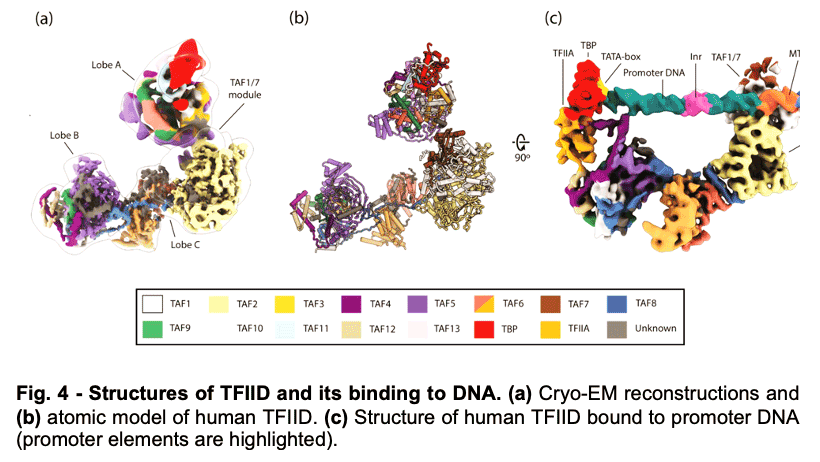 TFIID is a 1.3 MDa complex of TBP and 13 TBP associated factors (TAFs) that bind different core promoters, recruits the transcription pre-initiation complex (PIC), and integrates inputs from a variety of gene-specific transcription factors and epigenetic marks. We showed that human TFIID exhibits dramatic conformational rearrangements linked to its capacity to bind DNA, and we obtained the structures of the apo and DNA-bound TFIID (Fig. 4), allowing us to propose a model of how TFIID loads TBP for recruitment of a PIC only following the recognition of the core promoter. We proposed that TFIID binding to the core promoter involves the release of TBP from Lobe A in successive stages that overcome inhibitory interactions blocking different functional surfaces of TBP (Fig. 5).
TFIID is a 1.3 MDa complex of TBP and 13 TBP associated factors (TAFs) that bind different core promoters, recruits the transcription pre-initiation complex (PIC), and integrates inputs from a variety of gene-specific transcription factors and epigenetic marks. We showed that human TFIID exhibits dramatic conformational rearrangements linked to its capacity to bind DNA, and we obtained the structures of the apo and DNA-bound TFIID (Fig. 4), allowing us to propose a model of how TFIID loads TBP for recruitment of a PIC only following the recognition of the core promoter. We proposed that TFIID binding to the core promoter involves the release of TBP from Lobe A in successive stages that overcome inhibitory interactions blocking different functional surfaces of TBP (Fig. 5). 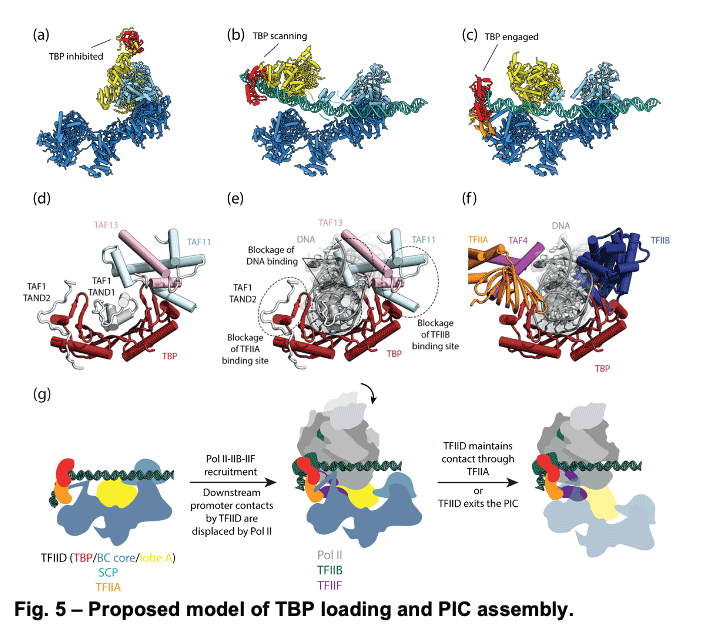 The interactions of the TAND1 and TAND2 regions of TAF1 with TBP 36 (Fig. 6a, d) are released (i) as Lobe C engages downstream DNA and positions the upstream promoter region such that it can be “scanned” by TBP (Fig. 5b, e), and (ii) as TFIIA joins the complex via interaction with Lobe B and further stabilizes the location of TBP for DNA interaction (Fig. 5c, f). The final engagement of TBP with the promoter is sterically incompatible with the TBP-TAF11 interaction and leads to TBP detachment from Lobe A and the opening of the binding site for TFIIB (Fig. 5c, f), which in turn will engage Pol II, promoting PIC assembly (Fig. 5g).
The interactions of the TAND1 and TAND2 regions of TAF1 with TBP 36 (Fig. 6a, d) are released (i) as Lobe C engages downstream DNA and positions the upstream promoter region such that it can be “scanned” by TBP (Fig. 5b, e), and (ii) as TFIIA joins the complex via interaction with Lobe B and further stabilizes the location of TBP for DNA interaction (Fig. 5c, f). The final engagement of TBP with the promoter is sterically incompatible with the TBP-TAF11 interaction and leads to TBP detachment from Lobe A and the opening of the binding site for TFIIB (Fig. 5c, f), which in turn will engage Pol II, promoting PIC assembly (Fig. 5g). -
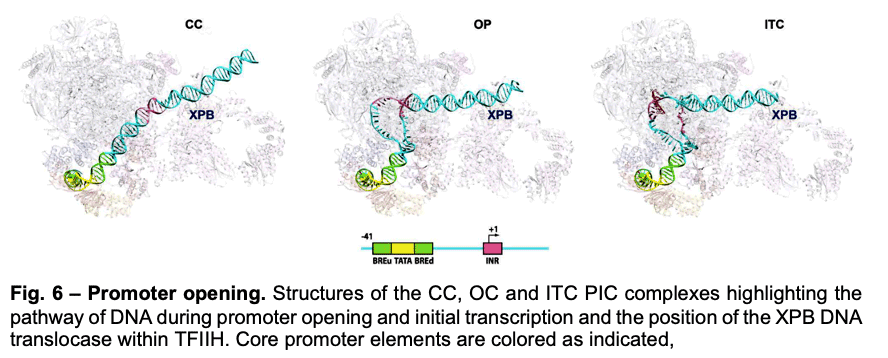
Deployment of TBP by TFIID upon core promoter recognition is followed by the assembly of the PIC around the transcription start site. We have described PIC structures (DNA-TBP-TFIIA-TFIIB-PolII-TFIIF-TFIIE-TFIIH) in three states throughout transcription initiation process: the initial close complex (CC), where the PIC engages duplex DNA; an open complex (OC), where the DNA contains an opened transcription bubble; and an initial transcribing complex (ITC), containing 6 RNA nucleotides. We showed that the TFIIB linker region is disordered in the CC state but becomes ordered upon interaction with the non-transcribed strand of the DNA, playing an essential role in stabilization of the transcription bubble (Fig.6). We found that the conformation of TFIIH changes upon PIC engagement Also, the position of XPB in the successive states of promoter engagement, bubble opening and initial transcription, agrees very well with a model in which this ATPase acts as a DNA translocase to move DNA into the active site of Pol II while generating torque that is release by unwinding of the DNA (Fig. 6).
Selected Publications
Greber, B.J., Remis, J., Ali, S. Nogales, E. (2021) Structure of the CDK-activating kinase bound to the clinical inhibitor ICE0942 at 2.5 Å resolution. Biophys. J. 120, 677-686 (journal cover).
Kasinath, V., Beck, C., Sauer, P., Poepsel, S., Kosmatka, J., Faini, M., Toso, D., Aebersold, R., and Nogales, E. (2021) JARID2 and AEBP2 regulate PRC2 activity in the presence of H2A ubiquitination or other histone modifications. Science 371, 6527.
Martin, R., Tiancong Qi, T., Zhang, H., Liu, F., King, M., Toth, C., Nogales, E and Staskawicz, B.J. (2020) Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ. Science 370, eabd 9993.
Patel, A.B., Moore, C.M., Greber, B.J., Luo, J., Zukin, S.A., Ranish, J. and Nogales, E. (2019) Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. Elife 54449.
Eshun-Wilson, L., Zhang, R., Portran, D., Nachury, M.V., Toso, D., Lohr, T., Vendruscolo, M., Bonomi, M., Fraser, J.S. and Nogales, E. (2019) Effects of α-tubulin acetylation on microtubule structure and stability. PNAS 116, 10366-10371.
Greber, B.J., Toso, D.B., Fang, J. and Nogales, E. (2019) The complete structure of the human TFIIH core complex. eLife 44771.
Liu, J-J. Orlova, N., Oakes, B.L., Ma, E., Spinner, H.B., Baney, K.L.M., Chuck, J., Tan, D., Knott, G.J., Harrington, L.B., Al-Shayeb, B., Wagner, A., Brötzmann, J., Staahl, B.T., Talyor, K.L., Desmarais, J., Nogales, E., Doudna, J.A. (2019) CRISPR-CasX is an RNA-dominated enzyme active for human genome editing. Nature 566, 218-223.
Patel, A. Louder, R.K., Greber, B.J., Grünberg, S., Luo, J., Fang, J., Liu, Y., Ranish, J., Hahn, S. and Nogales, E. (2018) Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872.
Kellogg, E.H., Hejab, N.M.A., Poepsel, S., Downing, K.H., DiMaio, F. and Nogales , E. (2018) Near-atomic model of microtubule-bound tau. Science 360, 1242-1246.
Nguyen, T.H.D., Tam, J., Wu, R.A., Greber, B.J., Toso, D., Nogales, E., Collins, K. (2018) Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 557, 190-195.
Pöpsel, S., Kasinath, V. and Nogales E. (2018) Cryo-EM structure of PRC2 simultaneous engagement with two functionally distinct nucleosomes. NSMB 25, 154-162
Kasinath, V., Faini, M., Reif, D., Feng, X.A., Stjepanovic, G., Poepsel, S., Aebersold, R. and Nogales, E. (2018) Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359, 940-944.
Zhang, R., Roostalu, J., Surrey, T. and Nogales, E. (2017) Structural Insight into TPX2-Stimulated Microtubule Assembly. eLife e30959.
Tenthorey, J.L., Haloupek, N., López- Blanco, J.R., Grob, P., Adamson, E., Hartenian, E, Lind, N.A., Chacón, P., Nogales, E and Vance, R.E. (2017) Structural basis of flagellin detection by NAIP5: a strategy to limit pathogen immune evasion. Science 358, 888-893.
Greber, B.J., Nguyen, T.H.D., Fang, J., Afonine, P.V., Adams, P.D. and Nogales, E. (2017) The cryo-EM structure of human TFIIH. Nature 549, 414-417.
Kellogg, E., Hejab, N.M.A., Howes, S., Northcote, P, Miller, J.H., Diaz, J.F., Downing, K.H. and Nogales, E. (2017). Insights into the distinct mechanisms of action of taxane and non-taxane microtubule stabilizers from cryo-EM studies. J. Mol. Biol. 429, 633–646.
He, Y., Yan, C., Inouye, C., Tjian, R., Ivanov, I. and Nogales, E. (2016) Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359-365..
Louder, R.K., He, Y. Lopez-Blanco, J.R. Fang, J., Chacon, P., and Nogales, E. (2016) Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604-609.
Jiang, F., Taylor, D.W., Chen, J.S., Kornfeld, J.E., Zhou, K., Thompson, A.W., Nogales, E. and Doudna, J.A. (2016) Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 35, 867-71.
Zhang, R., Alushin, G.M., Brown, A. and Nogales e. (2015) Mechanistic origin of microtubule dynamic instability and its regulation by EB proteins. Cell 162, 849-859.
Nogales, E. and Scheres, S.H.W. (2015) Cryo-EM: a unique tool for the visualization of molecular complexity. Mol. Cell 58, 677-689.
Alushin, G.M., Lander, G.C., Kellogg, E.H., Zhang, R., Baker, D. and Nogales, E. (2014) High- resolution microtubule structrues reveal the structural transitions in ab-tubulin upon GTP hydrolysis. Cell 157, 1117,1129. Preview in Cell; News and Views in NSMB (Jun 4).
Jinek, M., Jiang, F., Taylor, D.W., Sternberg, S.H., Kaya, E., Ma, E., Andres, C., Hauer, M., Zhou, K., Lin, S., Kaplan, M., Iavarone, A.T., Charpentier, E., Nogales, E. and Doudna, J.A. (2014) Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997.
Photo credit: Mark Hanson at Mark Joseph Studios.
Last Updated 2015-09-07