Gary Karpen

Professor of Cell Biology, Development and Physiology
Lab Homepage: https://sites.google.com/berkeley.edu/karpenlabResearch Interests
Our studies are focused on understanding inheritance, chromatin structure, gene expression, and the organization of chromosomes in the nucleus. Most of our studies have focused on the fruit fly Drosophila melanogaster as a model for chromosome function in metazoans, which allows us to address mechanisms in animals by synergistically combining molecular, genetic, cell biological and biochemical approaches. However, we have examined the relevance of our findings to human chromosomes, and have demonstrated surprising similarities between these evolutionarily-distant species.
Current Projects
We are currently pursuing three projects: 1) analysis of the determinants of centromere identity and function, 2) molecular-genetic dissection of proteins that regulate nuclear organization, and 3) sequence analysis of a poorly characterized genomic component, called heterochromatin. Recent results from the centromere project will be described here; access our lab web site for more details about this and other projects.
The centromere (CEN) is the chromosomal site associated with kinetochore formation, which is the structure responsible for microtubule attachment to the chromosome, and constitutes an essential component of prometaphase congression, mitotic checkpoint control, anaphase poleward segregation, and cytokinesis. Chromosome gain or loss (aneuploidy) results from having no kinetochore attachments, or multiple attachments. Aneuploidy has catastrophic effects on cells and organisms, and plays a key role in the surprisingly high frequency of human birth defects, and in cancer initiation and progression.
 How does the cell ensure that one, and only one region of the chromosome attaches to and moves along microtubules?? In other words, how is CEN identity propagated through replication and division? Significant evidence suggests that CEN identity is determined epigenetically, that is by mechanisms that function independent of the primary DNA sequence. Thus, chromatin structure, modification and replication are likely to play a critical role in CEN identity.
How does the cell ensure that one, and only one region of the chromosome attaches to and moves along microtubules?? In other words, how is CEN identity propagated through replication and division? Significant evidence suggests that CEN identity is determined epigenetically, that is by mechanisms that function independent of the primary DNA sequence. Thus, chromatin structure, modification and replication are likely to play a critical role in CEN identity.
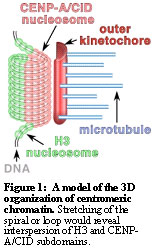
We have studied a constitutive component of centromeric chromatin (CENP-A), which substitutes for histone H3 in CEN nucleosomes. We have demonstrated that CENP-A is both a structural and a functional foundation for kinetochore formation. Three-dimensional deconvolution microscopy analysis demonstrated that CENP-A chromatin in flies and humans appears as a cylindrical structure, and that kinetochore proteins are wrapped around this structure. As expected from the composition of CEN nucleosomes, H3 appears to be excluded from the CENP-A cylinder. However, surprisingly, in analysis of two-dimensional chromatin preparations we observed that blocks of H3-containing chromatin are interspersed with CENP-A -containing chromatin.
How can we reconcile the uniformity of CENP-A and its separation from H3 in the higher order structure with the interspersion of H3 chromatin observed in two dimensions? We propose that the DNA may spiral (Figure 1) or loop through the cylinder, with CENP-A subdomains `stacked' into a cylinder, to the poleward side of a stack of interspersed H3 subdomains. We suggest that the function of this conserved structure may be to `present' centromeric chromatin to the external face of condensed mitotic chromosomes, where it is accessible to recruit kinetochore components and to attach to spindle microtubules.
RNA interference in tissue culture cells, live analysis of chromosome segregation after antibody injection into early embryos, and observations of flies with CENP-A null mutations demonstrated that CENP-A is required for all chromosome movements during mitosis and for normal cell cycle progression. Reciprocal epistasis experiments showed that CENP-A is very high in the kinetochore assembly pathway; CENP-A is required to recruit all known kinetochore proteins, but CENP-A localizes to centromeres in the absence of these proteins. We have also demonstrated that failure to form a kinetochore, due to depletion of CENP-A, results in the activation of a cell cycle checkpoint early in mitosis, prior to the time of a previously identified checkpoint known as the Spindle Assembly Checkpoint (SAC). Thus, CENP-A is physically and functionally positioned to be the epigenetic mark for centromere identity, or is closely associated with the mark.
If CENP-A is the epigenetic mark for propagation of centromere identity, how is new CENP-A deposited specifically at centromeres in response to replication and division? We hypothesize that `replenishment' in response to replication-generated segregation of CENP-A nucleosomes may be mediated by centromere-specific chromatin assembly complexes (CAFs) or remodeling factors. We are currently focused on in understanding the mechanisms and components responsible for specifically depositing CENP-A into CEN chromatin during or after replication. We have used biochemical and genetic approaches to identify CENP-A interacting proteins, and are determining if these candidates are required for CENP-A deposition.
Selected Publications
Sequence analysis of a functional Drosophila centromere. [X. Sun, H. Le, J. Wahlstrom, and G.H. Karpen (2003) Genome Research 13, 182-194]
Heterochromatic sequences in a Drosophila whole genome shotgun assembly. [R.A. Hoskins, et al. (2002) Genome Biology 3(12), RESEARCH0085]
Conserved organization of centromeric chromatin in flies and humans. [M.D. Blower, B.A. Sullivan, and G.H. Karpen (2002) Developmental Cell 2, 319-330]
The role of Drosophila CENP-A / CID in kinetochore formation, cell-cycle progression and interactions with heterochromatin. [M.D. Blower and G.H. Karpen (2001) Nature Cell Biology 3, 730-9]
The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. [K.L. Hari, K.R. Cook, G.H. Karpen (2001) Genes and Dev. 15, 1334-48]
Photo credit: Mark Hanson at Mark Joseph Studios.
Last Updated 2004-11-05