Current projects
Research
The control of T cell fate in the thymus
During thymic development, T cell precursors migrate to the thymus where they proliferate, rearrange their antigen receptor genes, and eventually give rise to the mature T cell subsets. During this process, the thymocytes are subject to a selection process that results in the death of ~99% of the cells and that shapes the mature T cell repertoire. We are investigating the mechanism that lead to positive and negative selection of T cells in the thymus, as well as the signaling events that control to cell fate decisions, including the CD4 versus CD8 cell lineage choice.
Immune responses to parasitic infection
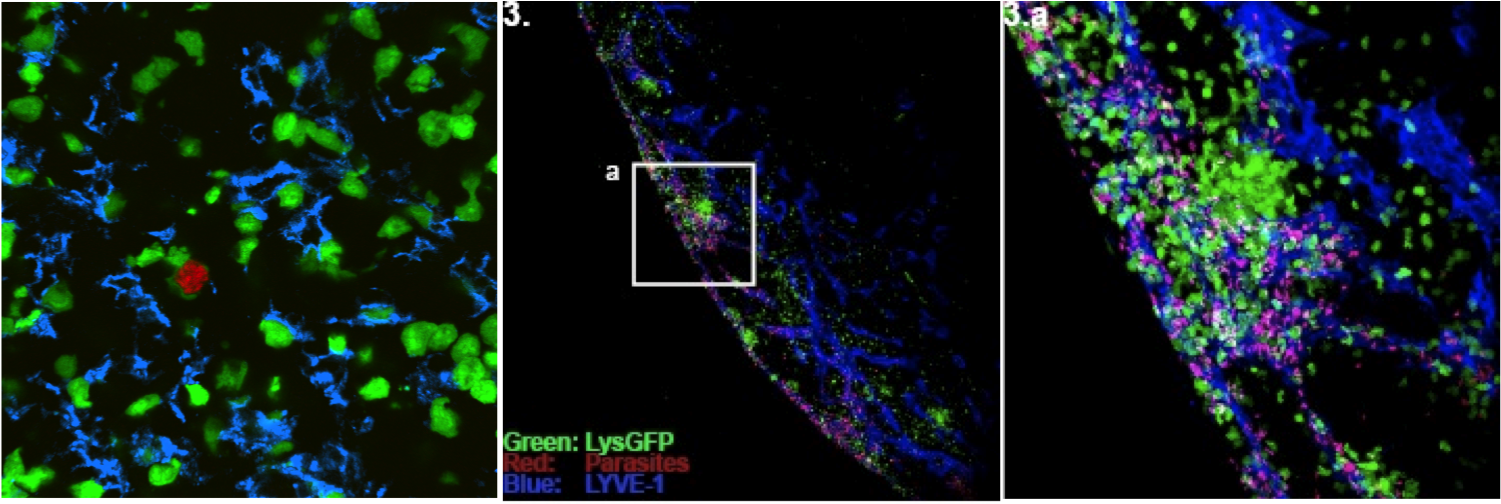
We are using mouse infection by the protozoan intracellular parasite, Toxoplasma gondii, as an experimental system for understanding the mammalian immune response. This model has particular relevance for understanding response to intracellular pathogens, and generating exhaustion-resistant CD8 T cell responses. We and our collaborators have identified the natural T cell antigens recognized in both resistant (Balb/c H-2d) and sensitive (C57Bl/6; H-2b) mouse strains, and are investigating the mechanisms that allow certain T cell responses to remain effective in the face of persistent infection.
Collaborative projects:
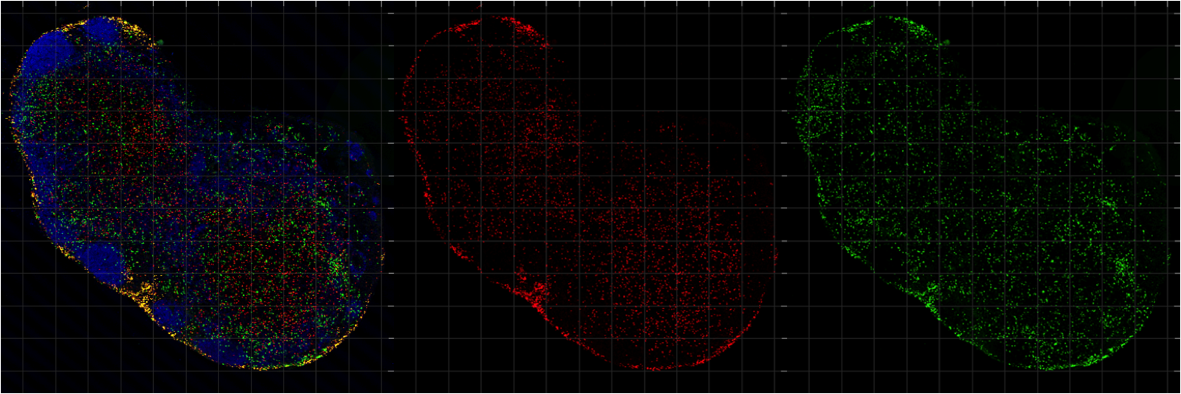
A temporal and spatial map of thymic development:To gain a deeper understanding of cell fate transitions in the thymus, we are engaged in a multidisciplinary collaborative project to generate a high resolution temporal and spatial map of thymic development with the labs of Nir Yosef https://niryosef.wordpress.com/group/and Aaron Streets http://streetslab.berkeley.edu/. We are combining single cell gene expression measurements (RNA and surface proteins), with spatial information about the location of developing T cells and support cells within the thymus, and with temporal information about changes in cell state over time. In the initial phase of the project, we have focused on the choice of developing T cells to develop into mature CD4 or CD8 T cells during positive selection. We have performed simultaneous measurement of mRNA and surface proteins (called TotalSeq or CiteSeq) on individual thymocytes, using samples from wild type mice, as well as mouse strains with biased cell fate decisions. This project extends and complements Human Cell Atlas projects, by providing a higher resolution analyses of key developmental branch points, and by leveraing the ability to perform in vivo and ex vivo experimental manipulations in mouse. 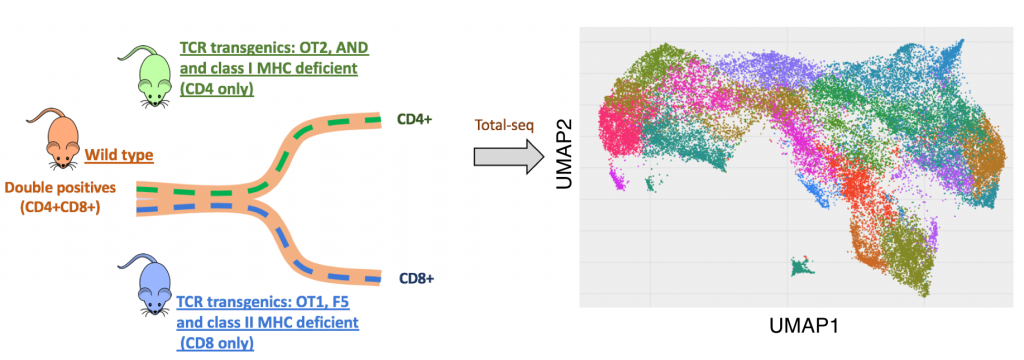

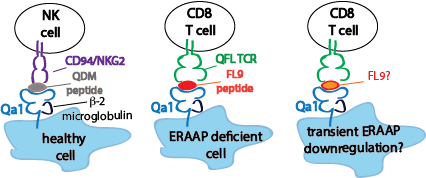
Thymic development of Qa1 restricted T cells: We are also investigating an interesting and poorly understood group of nonconventional T cells that recognize antigens associated with nonclassical MHC-1 molecules. Unlike classical MHC molecules, which are highly polymorphic, and play well defined roles in presenting pathogen derived antigens to T cells, nonclassical MHC molecules are relatively nonpolymorphic, bind a variety of antigens, including non-peptide antigens, and have relatively mysterious functions. Interestingly, the thymic development of T cells restricted to nonclassical MHC1 molecules appears to be quite distinct from those restricted to classical MHC, although this area remains understudied. Together with our collaborators, Nilabh Shastri http://labs.pathology.jhu.edu/shastri/ and Laurent Coscoy https://mcb.berkeley.edu/labs/coscoy/home, we have been investigating the thymic development of a group of CD8 T cells that recognize a self-peptide bound to the non classical MHC molecule Qa1
In healthy cells, the nonclassical MHC-1 molecule Qa1b presents the QDM peptide, inhibiting their killing by NK cells. When cells downregulate the MHC-1 antigen processing component ERAAP, Qa1b instead presents FL9 peptide exposing the cell to killing by QFL T cells.