ATP hydrolysis and substrate translocation
The 26S proteasome is composed of approximately 32 different subunits, forming the barrel-shaped 20S peptidase and the 19S regulatory particle. The 19S regulatory particle, also called the 19S cap, is an 18-subunit complex that functions in substrate recognition, processing, and as a gatekeeper for substrate entry to the 20S core. It consists of the lid and the base subcomplexes, the latter containing six distinct AAA+ ATPases that are probably arranged in a hexameric ring. Based on the homology with prokaryotic ATP-dependent unfoldases like ClpX, the base of the 19S cap is thought to mechanically unfold and translocate substrates in an ATP-hydrolysis dependent manner, but virtually nothing is known about the underlying molecular mechanisms.
A major goal of our research is to elucidate how many of the AAA+ subunits in the 19S base hydrolyze ATP or participate in substrate translocation, and what mechanisms are involved in subunit communication and coordination of activities in the hetero-hexameric ATPase ring. Furthermore, as there are no crystal structures available for the 19S complex or its individual ATPase subunits, the structural elements that face the central processing pore and contact the substrate polypeptide have yet to be determined. Our studies on the mechanisms of unfolding and translocation may also reveal determinants important for substrate “degradability” and give insight into how the structure, stability, and sequence properties of a protein affect its selection for proteolysis and the kinetics of degradation.
We are currently using the 26S proteasome from Saccharomyces cerevisiae, but will subsequently extend our studies to the mammalian system in vitro and in vivo. Given the role of the proteasome in the pathogenesis of numerous human diseases, a detailed knowledge of the molecular mechanisms of substrate processing may also lead to new ways of proteasome inhibition and thus novel pharmaceuticals for the treatment of cancer and other diseases.
Mechano-chemical coupling
We do not yet understand how AAA+ unfoldases transduce the energy from ATP binding/hydrolysis into conformational changes for substrate translocation. Our current model based on previous biochemical studies with the unfoldase ClpX from E. coli suggests that loop movement in the ATPase propel substrate through the central processing pore. We will use different spectroscopic methods to analyze the arrangement of structural elements within the 19S cap and their conformational changes upon ATP binding and hydrolysis. Besides monitoring conformational changes within the unfoldase, these measurements will also help to follow the path of substrate molecules during initial binding, engagement, unfolding, and translocation.
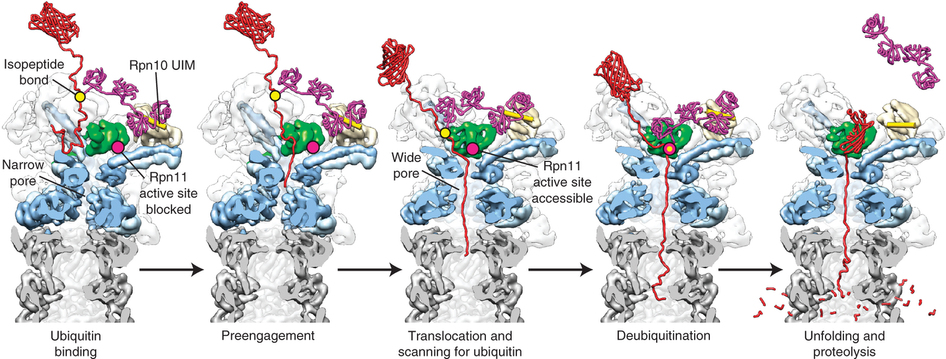
Substrate processing and de-ubiquitination
Another important challenge is to determine how the correct timing of substrate unfolding/translocation and de-ubiquitination influences degradation by the proteasome. The substrate is initially tethered to the 19S cap through its poly-ubiquitin modification, before a terminus or flexible structure gets engaged by the translocation machinery to initiate substrate processing. The poly-ubiquitin signal itself is cleaved off and recycled, allowing faster substrate degradation. However, if the poly-ubiquitin modification were released before proper engagement could occur, the substrate would lose its tether to the proteasome and escape proteolysis. Therefore, it seems likely that de-ubiquitination is coupled to engagement or translocation. The 19S cap includes several integral and associated de-ubiquitinating enzymes that cleave off or trim the poly-ubiquitin chain, as well as ubiquitin ligases that catalyze chain extension. Hence, a complex system of linked de-ubiquitination/re-ubiquitination and translocation reactions may act as a timer to regulate substrate degradation. The rate of engagement and unfolding by the 19S complex relative to the rate of trimming the poly-ubiquitin chain is probably a critical factor in determining the fate of a particular substrate.