2025
MacRae NS, Dong KC, Harimoto H, Martin A. Identification of novel ubiquitin receptors on the 26S proteasome by photo-crosslinking mass spectrometry. (2025) bioRxiv doi: 10.64898/2025.12.19.695558 PMID: 41446218; PMCID: PMC12723993.
Liao Z, Arkinson C, Martin A. Faf1 accelerates p97-mediated protein unfolding by promoting ubiquitin engagement. (2025) bioRxiv doi: 10.1101/2025.10.27.684972 PMID: 41278724; PMCID: PMC12636405.
Arkinson C, Gee CL, Zhang Z, Dong KC, Martin A. Structural landscape of the degrading 26S proteasome reveals conformation-specific binding of TXNL1. (2025) Nature Struct Mol Biol doi: 10.1038/s41594-025-01695-2 PMID: 41198955; PMCID:
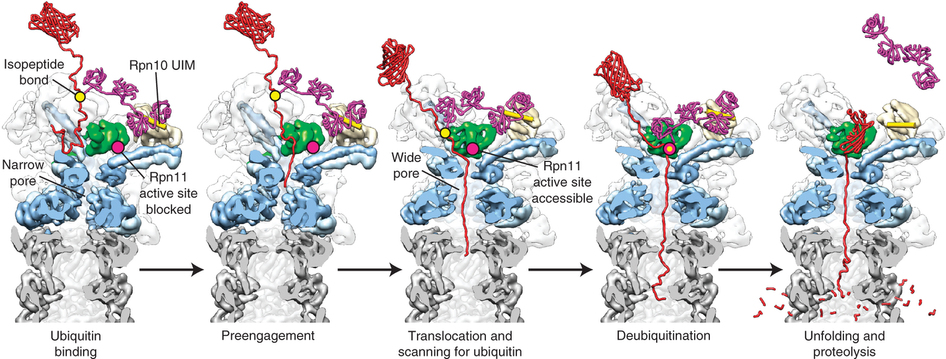
Htet ZM, Dong KC, Martin A. The deubiquitinase Rpn11 functions as an allosteric ubiquitin sensor to promote substrate engagement by the 26S proteasome. (2025) Cell Reports 44(6):115736 doi: 10.1016/j.celrep.2025.115736 PMID: 40411784; PMCID: PMC12256112
Arkinson C, Dong KC, Gee CL, Costello SM, Soe AC, Hura GL, Marqusee S, Martin A. NUB1 traps unfolded FAT10 for ubiquitin-independent degradation by the 26S proteasome. (2025) Nat Struct Mol Biol 32(9):1752-1765 doi: 10.1038/s41594-025-01527-3. PMID: 40217121
Arkinson C, Dong KC, Gee CL, Martin A. Mechanisms and regulation of substrate degradation by the 26S proteasome. (2025) Nat Rev Mol Cell Biol26(2):104-122
doi: 10.1038/s41580-024-00778-0 PMID: 39362999; PMCID: PMC11772106
2024
Yori Restrepo S, Martin A. Recombinant Expression of Photo-crosslinkable 26S Proteasome Base Subcomplex. (2024) bioRxiv
doi: https://doi.org/10.1101/2024.12.16.628829
Sascha J. Amann, Ken Dong, Josef Roehsner, Dominik Krall, Irina Grishkovskaya, Harald Kotisch, Alexander Schleiffer, Elisabeth Roitinger, Andrea Pauli, Andreas Martin, David Haselbach.PITHD1: An Endogenous Inhibitor of the 26S Proteasome During Cellular Dormancy. (2024) bioRxiv
doi: https://doi.org/10.1101/2024.12.04.626795
Arkinson C, Gee CL, Zhang Z, Dong KC, Martin A. Structural landscape of AAA+ ATPase motor states in the substrate-degrading human 26S proteasome reveals conformation-specific binding of TXNL1. (2024) bioRxiv
doi:10.1101/2024.11.08.622731 PMID: 39574680; PMCID: PMC11580985
Htet ZM, Dong KC, Martin A. The deubiquitinase Rpn11 functions as an allosteric ubiquitin sensor to promote substrate engagement by the 26S proteasome. (2024) bioRxiv
doi: 10.1101/2024.10.24.620116 PMID: 39484543; PMCID: PMC11527175.
Arkinson C, Dong KC, Gee CL, Costello SM, Marqusee S, Martin A. Nub1 traps unfolded FAT10 for ubiquitin-independent degradation by the 26S proteasome. (2024)
bioRxiv.
doi: 10.1101/2024.06.12.598715 PMID: 38915702
2023
López-Alfonzo EM, Saurabh A, Zarafshan S, Pressé S, Martin A. Substrate-interacting pore loops of two ATPase subunits determine the degradation efficiency of the 26S proteasome. (2023) bioRxiv
doi: https://doi.org/10.1101/2023.12.14.571752
Williams C, Dong KC, Arkinson C, Martin A. Preparation of site-specifically fluorophore-labeled polyubiquitin chains for FRET studies of Cdc48 substrate processing. (2023) STAR Protoc 4(4):102659.
doi: 10.1016/j.xpro.2023.102659 PMID: 37889757; PMCID: PMC10630674.
Williams C, Dong KC, Arkinson C, Martin A. The Ufd1 cofactor determines the linkage specificity of polyubiquitin chain engagement by the AAA+ ATPase Cdc48. (2023) Mol Cell 83(5):759-769.e7.
doi: 10.1016/j.molcel.2023.01.016 PMID: 36736315; PMCID: PMC9992269.

Xie G, Dong KC, Worden EJ, Martin A. High-Throughput Assay for Characterizing Rpn11 Deubiquitinase Activity. (2023) Methods Mol Biol 2591:79-100.
doi: 10.1007/978-1-0716-2803-4_6. PMID: 36350544.
2022
Jonsson E, Htet ZM, Bard JAM, Dong KC, Martin A. Ubiquitin modulates 26S proteasome conformational dynamics and promotes substrate degradation. (2022) Sci Adv 8(51):eadd9520.
doi: 10.1126/sciadv.add9520 PMID: 36563145; PMCID: PMC9788759.
2021
Jonsson E, Htet ZH, Bard JAM, Dong K & Martin A. Ubiquitin modulates 26S proteasome conformational dynamics and promotes substrate degradation. (2021) bioRxiv
doi:10.1101/2021.08.18.456915
Didychuk AL, Gates SN, Gardner MR, Strong LM, Martin A, Glaunsinger BA. A pentameric protein ring with novel architecture is required for herpesviral packaging. (2021) Elife 10:e62261.
doi: 10.7554/eLife.62261 PMID: 33554858; PMCID: PMC7889075.
Chen X, Htet ZM, López-Alfonzo E, Martin A, Walters KJ. Proteasome interaction with ubiquitinated substrates: from mechanisms to therapies. (2021) FEBS J 288(18):5231-5251.
doi:10.1111/febs.15638 PMID: 33211406; PMCID: PMC8131406.
2020
Castanzo DT, LaFrance B & Martin A. The AAA+ ATPase Msp1 is a processive protein translocase with robust unfoldase activity. (2020) Proc Natl Acad Sci USA 117(26):14970-14977.
doi: 10.1073/pnas.1920109117
Carroll EC, Greene ER, Martin A & Marqusee SM. Site-specific ubiquitination affects protein energetics and proteasomal degradation. (2020) Nature Chemical Biology 16:866–875
doi: 10.1038/s41589-020-0556-3
[News and Views: Unraveling proteasome engagement]
Greene ER, Dong KC & Martin A. Understanding the 26S proteasome molecular machine from a structural and conformational dynamics perspective. (2020) Curr Opin Struct Biol 61:33-41.
doi: 10.1016/j.sbi.2019.10.004
2019
Greene ER, Goodall EA, de la Peña AH, Matyskela ME, Lander GC & Martin A. Specific lid-base contacts in the 26s proteasome control the conformational switching required for substrate degradation. (2019) eLife 8:e49806.
doi: 10.7554/eLife.49806
Gates SN & Martin A. Stairway to translocation: AAA+ motor structures reveal the mechanisms of ATP‐dependent substrate translocation. (2019) Protein Science 29(2):407-419.
doi: 10.1002/pro.3743
Blythe E.E., Gates S.N., Deshaies R.J., Martin A. Multisystem Proteinopathy Mutations in VCP/p97 Increase NPLOC4·UFD1L Binding and Substrate Processing. (2019) Structure 27(12):1820-1829.e4.
doi: 10.1016/j.str.2019.09.011 PMID: 31623962; PMCID: PMC6929323.
Bard, J.A.M., Bashore, C., Dong, K.C. & Martin, A. The 26S Proteasome Utilizes a Kinetic Gateway to Prioritize Substrate Degradation. Cell 177:1-13 (2019).
doi:10.1016.cell.2019.02.031
Olszewski, M. M., Williams, C., Dong, K. C. & Martin, A. The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome. (2019) Communications Biology 2:29.
doi: 10.1038/s42003-019-0283-z
2018
de la Peña AH*, Goodall EA*, Gates SN*, Lander GC, Martin A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis–driven translocation. (2018) Science 362:6418.
doi: 10.1126/science.aav0725
Bard JAM & Martin A. (2018) Recombinant Expression, Unnatural Amino Acid Incorporation, and Site-Specific Labeling of 26S Proteasomal Subcomplexes. In: Mayor T., Kleiger G. (eds) The Ubiquitin Proteasome System. Methods in Molecular Biology, vol. 1844. Humana Press, New York, NY.
doi: 10.1007/978-1-4939-8706-1_15
Bard JAM*, Goodall EA*, Greene ER, Jonsson E, Dong KC, Martin A. Structure and Function of the 26S Proteasome. (2018) Annu Rev Biochem 87:697-724.
doi: 10.1146/annurev-biochem-062917-011931
Gardner BM, Castanzo DT, Chowdhury S, Stjepanovic G, Stefely MS, Hurley JH, Lander GC, Martin A. The peroxisomal AAA-ATPase Pex1/Pex6 unfolds substrates by processive threading. (2018) Nat Commun 9:135.
doi: 10.1038/s41467-017-02474-4
2017
San Martín Á, Rodriguez-Aliaga P, Molina JA, Martin A, Bustamante C & Baez M. Knots can impair protein degradation by ATP-dependent proteases. (2017) Proc Natl Acad Sci USA 114:9864–9869.
doi: 10.1073/pnas.1705916114
Worden EJ, Dong KC & Martin A. An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome. (2017) Mol Cell 67:1–22.
doi: 10.1016/j.molcel.2017.07.023
2016
Rodriguez-Aliaga P, Ramirez L, Kim F, Bustamante C & Martin A. Substrate-translocating loops regulate mechanochemical coupling and power production in AAA+ protease ClpXP. (2016) Nat Struct Mol Biol 23:974–981.
doi: 10.1038/nsmb.3298
Dambacher CM*, Worden EJ*, Herzik MA, Martin A & Lander GC. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. (2016) eLife 5:1-17.
doi: 10.7554/eLife.13027
2015
Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC & Martin A. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. (2015) Nat Struct Mol Biol 22:712–719.
doi: 10.1038/nsmb.3075
Yang B, Stjepanovic G, Shen Q, Martin A & Hurley JH. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. (2015) Nat Struct Mol Biol 22:492–498
doi: 10.1038/nsmb.3015
Gardner BM, Chowdhury S, Lander GC, Martin A. The Pex1/Pex6 Complex Is a Heterohexameric AAA+ Motor with Alternating and Highly Coordinated Subunits. (2015) J Mol Biol 427:1375-1388.
doi: 10.1016/j.jmb.2015.01.019 PMID: 25659908; PMCID: PMC4355278.
2014
Worden EJ, Padovani C & Martin A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. (2014) Nat Struct Mol Biol 21:220–227.
doi: 10.1038/nsmb.2771 PMID: 24463465.
Nyquist, K. & Martin, A. Marching to the beat of the ring: polypeptide translocation by AAA+ proteases. (2014) Trends Biochem Sci 39, 53-60.
10.1016/j.tibs.2013.11.003 PMID: 24316303; PMCID: PMC3946816.
2013
Sen M*, Maillard RA*, Nyquist K*, Rodriguez-Aliaga P, Pressé S, Martin A & Bustamante C. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. (2013) Cell 155:636-46.
doi: 10.1016/j.cell.2013.09.022 PMID: 24243020; PMCID: PMC3901371.
Beckwith R, Estrin E, Worden EJ & Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. (2013) Nat Struct Mol Biol 10:1164-72. doi: 10.1038/nsmb.2659 PMID: 24013205; PMCID: PMC3869383.

Estrin E, Lopez-Blanco JR, Chacon P & Martin A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. (2013) Structure 21:1-12.
doi: 10.1016/j.str.2013.06.023 PMID: 23911091.
Matyskiela ME, Lander GC & Martin A. Conformational switching of the 26S proteasome enables substrate degradation. (2013) Nat Struct Mol Biol 20:781-8.
doi: 10.1038/nsmb.2616 PMID: 23770819; PMCID: PMC3712289.
Lander GC, Martin A & Nogales E. The proteasome under the microscope: the regulatory particle in focus. (2013) Curr Opin Struct Biol 23:243-51.
doi: 10.1016/j.sbi.2013.02.004PMID: 23498601; PMCID: PMC3676703.
Matyskiela ME & Martin A. Design principles of a universal protein degradation machine. (2013) J Mol Biol 425:199-213.
doi: 10.1016/j.jmb.2012.11.001 PMID: 23147216; PMCID: PMC3546117.
2012
Lander GC*, Estrin E*, Matyskiela ME*, Bashore C, Nogales E & Martin A. Complete subunit architecture of the proteasome regulatory particle. (2012) Nature 482:186-91.
doi: 10.1038/nature10774 PMID: 22237024; PMCID: PMC3285539.
2011
Maillard RA, Chistol G, Sen M, Righini M, Tan J, Kaiser CM, Hodges C, Martin A & Bustamante C. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. (2011) Cell 145:459-69.
doi: 10.1016/j.cell.2011.04.010 PMID: 21529717; PMCID: PMC3686100.
2009
Glynn, SE, Martin A, Nagar AR, Baker TA & Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. (2009) Cell 139:744-56.
doi: 10.1016/j.cell.2009.09.034 PMID: 19914167; PMCID: PMC2778613.
2008
Martin A, Baker TA & Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. (2008) Nat Struct Mol Biol 15:1147-51.
doi: 10.1038/nsmb.1503 PMID: 18931677; PMCID: PMC2610342.
Martin A, Baker TA & Sauer RT. Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. (2008) Nat Struct Mol Biol 15, 139-45.
doi: 10.1038/nsmb.1380 PMID: 18223658.
Martin A, Baker TA & Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. (2008) Mol. Cell 29:441-50.
doi: 10.1016/j.molcel.2008.02.002 PMID: 18313382; PMCID: PMC2323458.
2007
Martin A, Baker TA & Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. (2007) Mol Cell 27:41-52. doi: 10.1016/j.molcel.2007.05.024 PMID: 17612489; PMCID: PMC2074893.
2005
Martin A, Baker TA & Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. (2005) Nature 437:1115-20.
doi: 10.1038/nature04031 PMID: 16237435.