We are interested in understanding quantitatively the effects of single and multiple amino acid substitutions on: 1. enzyme activity, 2. protein stability and 3. protein-protein complexes. Our recently introduced formal analysis of chimeric constructs (Luong and Kirsch 2001) is proving applicable to the above areas of interest and has the potential to be useful in protein design applications. Current projects are listed below:
 References
ReferencesWe have used high resolution epitope mapping to understand how specific contacts at interfaces effect both the stability and kinetics of these interactions. We recently devised an experimental approach that reveals details about docking trajectories of interacting proteins. (Taylor et al, 1998). This method is being adapted to evaluate subunit associations in oligomeric proteins. Protein stability and protein-protein interactions are being examined by mass spectrometry in collaboration with Professor E. Williams of this university.
Selected Protein-Protein Recognition Publications
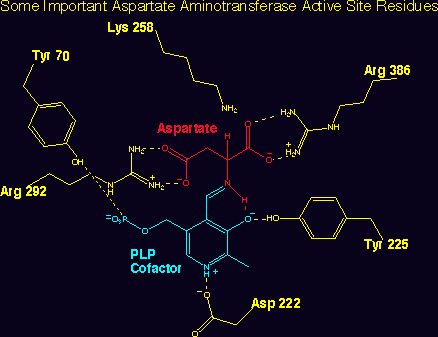
The laboratory has a long standing interest in the biochemistry of enzymes that utilize the cofactor pyridoxal phosphate (PLP), which is essential for the catalysis of the major reactions involved in amino acid transformations. Projects 1-5 illustrate some of our current pursuits in PLP enzymology: 1) The best understood of these enzymes is aspartate aminotransfersase. The three- dimensional structure of this enzyme has guided these experiments, and has aided greatly in the interpretation of the mechanistic studies carried out in our laboratory. We are currently employing directed evolution by DNA shuffling to evolve the selectivity of aspartate aminotransferase, which acts selectively on aspartate and glutamate, to an aromatic amino acid aminotransferase, which reacts preferentially with phenylalanine and tyrosine.
Selected Aminotransferase Publications
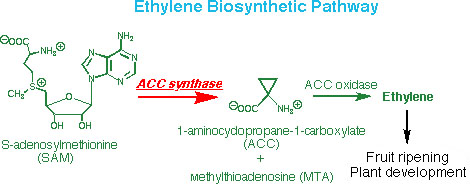
2) The enzyme aminocyclopropane carboxylate synthase (ACCS) is the committed enzyme in the biosynthetic pathway to the important plant hormone, ethylene. Our laboratory, in a collaboration, has solved the crystal structure, and we are currently involved in a detailed study of the mechanism of action of this enzyme.
![]() Selected ACC Synthase Publications
Selected ACC Synthase Publications
3) We are collaborating with the group of Gunther Schneider of the Karolinska Institute in Stockholm on an unusual aminotransferase employing S-adenosyl methionine as the amino donor in the biosynthetic pathway of biotin.
4) The enzyme cystathionine ß-synthase (CBS) is connected with a variety of human diseases derived from its malfunction. The defect results in homocystinuria, and nearly 100 disease associated mutations in CBS have been documented. We are attempting to discover the role of the specific mutations involved in the etiology of the diseases.
5) The recently completed genome sequences for a number of organisms have led us to discover a number of previously undescribed ORFs that encode for pyridoxal phosphate binding proteins of unknown activity. The functions are being pursued by biochemical analysis of the expressed enzymes together with genomics and proteomics approaches.
1) Venn divides the residues of a sequence into sets based on their conservation in groups of sequences that the user defines. For example, sequences might be grouped by the preferred substrate of the enzyme they encode and conserved residues identified by Venn for each substrate specificity. The output can be represented as a Venn Diagram. See Rothman and Kirsch (2003) and Deu, et al (2002) for implementations.
- Download the lastest Linux version here (accepts multiple sequence alignments in FASTA format as input).
2) Patterns looks for covariance of residues with function. It is especially useful for higher-order covariation involving more than 2 positions.
- Download the latest Windows version here and the Read Me here.
 Publications
Publications[Home] [The Boss] [Research] [Publications] [Links] [The Group] [For Students]
|
Departments of Molecular and Cell Biology and Chemistry University of California, Berkeley and Materials Sciences Division Lawrence Berkeley National Laboratory Berkeley, California 94720 +1-510-642-6368 |
copyright 1996 University of California
page maintained by |