Molecular Networks Controlling Dynamic
Chromosome Behaviors during Development
Our research focuses on the interplay
between the structure of chromosomes and
their function. Chromosomes undergo
dynamic behaviors during development to
ensure genome stability and accurate cell fate
decisions. We study inter-related molecular
networks that control diverse chromosome
behaviors: chromosome counting to
determine sexual fate; X-chromosome
remodeling to achieve X-chromosome
repression during dosage compensation, an
epigenetic process; chromosome cohesion to
tether and release replicated chromosomes
for reducing genome copy number during
germ cell formation; and chromosome
compaction to control gene expression,
chromosome segregation, and recombination
between maternal and paternal
chromosomes. We have found that the
developmental control of gene expression is
achieved through chromatin modifications
that affect chromosome structure with
epigenetic consequences. We have also
established robust procedures for targeted
genome editing across nematode species
diverged by 300 MYR to study the evolution of
sex determination and dosage compensation.
We combine genetic, genomic, proteomic,
biochemical, and cell biological approaches to
study these questions in the model organism
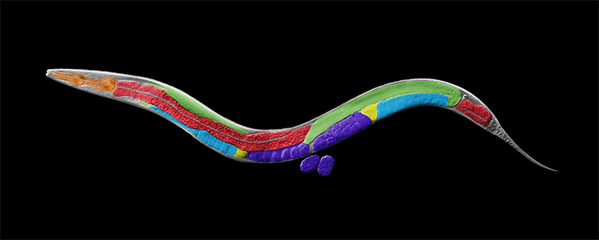
Caenorhabditis elegans, a round worm, and
its related nematode species.
Counting Chromosomes to Determine Sex:
Molecular Antagonism between
X-Chromosome, Autosome Signals Specifies
Nematode Sex
Many organisms determine sexual fate by a
chromosome-counting mechanism that
distinguishes one X chromosome from two.
Embryos with one X become males, while
those with two become females. We dissected
the molecular mechanism by which the
nematode C. elegans counts its sex
chromosomes to discern how small changes
in the concentrations of molecular signals are
translated into dramatically different
developmental fates. C. elegans tallies
X-chromosome number relative to the ploidy,
the sets of autosomes (X:A signal). It
discriminates with high fidelity between
tiny differences in the signal: 2X:3A embryos
(ratio 0.67) become males, while 3X:4A
embryos (ratio 0.75) become hermaphrodites
(Figure 1).
We showed that a set of X-linked genes called
X-signal elements (XSEs) communicates
X-chromosome dose by repressing the master
sex-determination switch gene xol-1 in a
cumulative, dose-dependent manner. XOL-1, a
GHMP kinase, is activated in 1X:2A embryos (1
dose of XSEs) to set the male fate but
repressed in 2X:2A embryos (2 doses of XSEs)
to promote the hermaphrodite fate, including
the activation of X-chromosome dosage
compensation. We also showed that the dose
of autosomes is communicated by a set of
autosomal signal elements (ASEs) that also act
in a cumulative, dose-dependent manner to
counter XSEs by stimulating xol-1
transcription. We have explored the
biochemical basis by which XSEs counter ASEs
to determine sex. Analysis in vitro showed
that XSEs (nuclear receptors and
homeodomain proteins) and ASEs (T-box and
zinc-finger proteins) bind directly to at least 5
distinct sites in xol-1 regulatory DNA to
counteract each other's activities and thereby
regulate xol-1 transcription (Figure 2). Analysis
in vivo showed that disrupting ASE and XSE
binding sites recapitulated the mis-regulation
of xol-1 transcription caused by disrupting the
cognate signal element genes. XSE and ASE
binding sites are distinct and non-overlapping,
suggesting that direct competition for xol-1
binding is not the mechanism by which XSEs
counter ASEs. Instead, XSEs likely antagonize
ASEs by recruiting cofactors with reciprocal
activities that induce opposite transcriptional
states. The X:A balance is thus communicated
in part through multiple antagonistic
molecular interactions carried out on a single
promoter, revealing how small differences in
X:A values can elicit different sexual fates. We
are currently identifying potential coactivators
and corepressors and directing efforts toward
understanding the evolution of the X:A signal
across nematode species.
Although most XSEs repress xol-1 by
regulating transcription, one XSE, an RNA
binding protein, represses xol-1 by binding to
an alternatively spliced intron and blocking its
proper splicing, thereby generating a
non-functional transcript with an in-frame
stop codon (Figure 2). This second tier of
repression enhances the fidelity of the
counting process.
The concept of a sex signal comprising
competing XSEs and ASEs arose as a theory
for fruit flies one century ago, and it
subsequently became entrenched in
textbooks. Ironically, the recent work of others
showed the fly sex signal does not fit this
simple paradigm, but our work shows the
worm signal does.
X-Chromosome Dosage Compensation:
Repressing X Chromosomes via Molecular
Machines.
Organisms that use sex chromosomes to
determine sexual fate evolved the essential,
chromosome-wide regulatory process called
dosage compensation to balance
X-chromosome gene expression between the
sexes. Strategies for dosage compensation
differ from worms to mammals, but invariably
a regulatory complex is targeted to X
chromosomes of one sex to modulate
transcription along the entire chromosome.
The heritable, regulation of X-chromosome
expression during dosage compensation is
exemplary for dissecting the coordinate
regulation of gene expression over large
chromosomal territories and the role of
chromosome structure in regulating gene
expression.
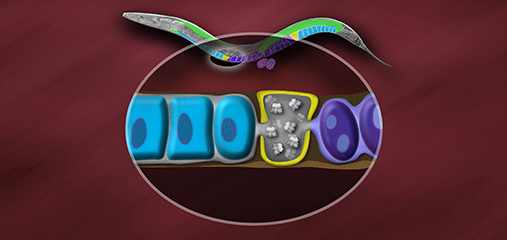
We defined the C. elegans dosage
compensation complex (DCC) and showed it is
homologous to condensin, a conserved
protein complex that mediates the
compaction, resolution, and segregation of
mitotic and meiotic chromosomes from yeast
to humans (Figure 3). The DCC binds to both X
chromosomes of hermaphrodites to reduce
transcription by half (Figure 3). Failure to
reduce expression kills hermaphrodites. Most
DCC condensin subunits also control the
structure and function of mitotic and meiotic
chromosomes by participating in two other
distinct condensin complexes (Figure 3). Not
only has the DCC co-opt subunits of
condensin to control gene expression, it
co-opted a subunit from the MLL/COMPASS
complex, a histone modifying complex, to
help recruit condensin subunits to rex sites.
We found that the DCC condensin subunits
are recruited specifically to hermaphrodite X
chromosomes by sex-specific DCC subunits
that trigger binding to cis-acting regulatory
elements on X, called rex and dox sites. rex
(recruitment elements on X) sites recruit the
DCC in an autonomous, sequence-dependent
manner using DNA motifs highly enriched on
X chromosomes. The DCC spreads to dox
(dependent on X) sites, which reside in
promoters of active genes and bind the DCC
robustly only when linked to rex sites.
Dynamic Control of X-Chromosome
Conformation and Repression by a Histone
H4K20me Demethylase.
We recently found that DCC subunit DPY-21
has a histone demethylase activity that is
responsible for the selective enrichment of
H4K20me1 on X chromosomes of XX embryos
upon DCC binding. X-ray crystallography and
biochemical assays of DPY-21 revealed a novel
subfamily of Jumonji C histone demethylases
that converts H4K20me2 to H4K20me1.
Selective inactivation of demethylase activity
in vivo by genome editing eliminated
H4K20me1 enrichment on X, elevated X-linked
gene expression, reduced X-chromosome
compaction, and disrupted X-chromosome
topology by weakening TAD boundaries.
These findings, among others, demonstrate
the direct impact of chromatin modification
on higher-order chromosome structure in
long-range regulation of gene expression.
H4K20me1 is also enriched on the
mammalian inactive X chromosome, but the
role of this enrichment in mammalian
transcriptional silencing is not known, nor is a
selective reagent available to test its role. We
showed that the mouse homolog of the DCC
subunit also has H4K20me2 demethylase
activity. Hence the worm system holds great
promise for understanding the effects of
histone modification in mammals.
Unexpectedly, DPY-21 associates with
autosomes but not X chromosomes of germ
cells in a DCC-independent manner to enrich
H4K20me1 and facilitate chromosome
compaction. Thus, DPY-21 is an adaptable
chromatin regulator that is harnessed during
development for distinct biological functions.
In both somatic cells and germ cells,
H4K20me1 enrichment modulates 3D
chromosome architecture to carry out these
functions.
Step of transcription controlled by the DCC.
We have dissected a key aspect of the dosage
compensation mechanism by determining the
step of transcription controlled by the DCC to
repress X-chromosome gene expression. This
work was performed in collaboration with
John Lis' lab at Cornell University. In principle,
the DCC could control any step of
transcription: recruitment of RNA polymerase
II (Pol II) to the promoter, initiation of
transcription, escape of Pol II from the
promoter or pause sites, elongation of RNA
transcripts, or termination of transcription.
The mechanism had been elusive in C. elegans
due to improper annotation of transcription
start sites (TSSs). Nascent RNA transcripts
from most nematode genes undergo rapid
co-transcriptional processing in which the 5'
end is replaced by a common 22-nucleotide
leader RNA through a trans-splicing
mechanism, thereby destroying all knowledge
of TSSs and promoters.
To understand the step of transcription
controlled by the DCC, we first devised a
general strategy for mapping transcription
start sites and created an invaluable
nematode TSS data set. The TSS mapping
strategy, called GRO-cap, recovered nascent
RNAs with 5'-caps prior to processing. We
then determined the genome-wide
distribution, orientation, and quantity of
transcriptionally-engaged RNA Polymerase II
(Pol II) relative to TSSs in wild-type and
DC-defective animals using GRO-seq (global
run-on sequencing).
We found that promoters are unexpectedly
far upstream from the 5' ends of mature
mRNAs, and promoter-proximal Pol II pausing
occurs only in starved larvae and is rare in C.
elegans embryos, unlike in most metazoans.
These results indicated that enhancement of
promoter pausing in XX embryos cannot be
the mechanism of reducing transcription
during dosage compensation. In contrast,
control of pausing is a common mechanism
for controlling transcription of developmental
regulatory genes in most metazoans and is
thought to be the mechanism of dosage
compensation in fruit flies.
Then, by comparing the location and density
of transcriptionally engaged Pol II in wild-type
and dosage-compensation-defective embryos,
we found that the step of transcription
controlled by the dosage compensation
process is the recruitment of Pol II. That is, C.
elegans equalizes X-chromosome-wide gene
expression between the sexes by reducing Pol
II recruitment to the promoters of X-linked
genes in XX embryos by about half. One of
our research directions is to dissect the
mechanisms by which the DCC limits Pol II
recruitment.
Our data set also enabled us to analyze
starvation-controlled gene regulation in
collaboration with Ryan Baugh's lab at Duke
University. We found a new phenomenon of
Pol II docking, the stable association of Pol II
upstream of the transcription start sites, and
hence sites of pausing. We found that docked
Pol II accumulates, without initiating,
upstream of inactive growth genes that are
turned off during starvation are activated
upon feeding. We found that Pol II pausing
occurs at active stress-response genes that
are downregulated upon feeding. Hence,
growth and stress genes are controlled by
distinct mechanisms to coordinate gene
expression with nutrient availability.
DCC recruitment and binding to X
chromosomes.
We showed that many of the DCC recruitment
(rex) sites have a DNA motif (called MEX) that
is highly enriched on X compared to
autosomes and is essential for DCC binding to
a subset of rex sites. However, not all rex sites
have this motif. We recently defined new
principles by which the DCC is recruited to X
chromosomes, including the identification of a
new, essential DCC binding motif (MEX II) that
is enriched on X. We found that MEX II acts in
combination with MEX to foster high-affinity
binding at some rex sites but also acts alone at
other rex sites to foster stable binding. We
demonstrated these DCC binding principles by
using DCC binding assays in vivo and in vitro.
We also showed that SUMOylation of specific
DCC subunits is essential for sex-specific
assembly and function of the DCC on X.
Depletion of SUMO in vivo severely disrupts
DCC binding and causes changes in X-linked
gene expression similar to those caused by
deleting the genes that encode DCC subunits.
Three DCC subunits undergo SUMOylation,
one subunit essential for DCC loading and two
subunits that are integral to the condensin
portion of the DCC.
DCC SUMOylation is triggered by the signal
that initiates DCC assembly onto X. The initial
step of assembly--binding of X-targeting
factors to rex sites--is independent of
SUMOylation, but robust binding of the
complete complex requires SUMOylation. One
of SUMOylated DCC subunits also participates
in condensin complexes essential for
chromosome segregation, but its
SUMOylation occurs only in the context of the
DCC. Our results reinforce a newly emerging
theme in which multiple proteins of a complex
are collectively SUMOylated in response to a
specific stimulus, leading to accelerated
complex formation and enhanced function.
Condensin-driven remodeling of
X-chromosome topology during dosage
compensation.
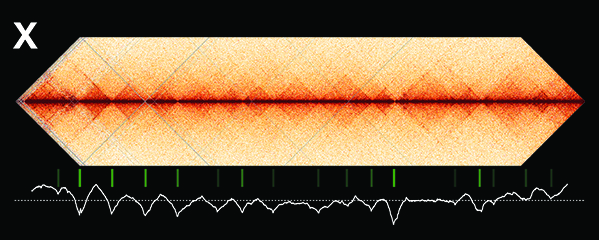
The three-dimensional organization of a
genome plays a critical role in regulating gene
expression, yet little is known about the
machinery and mechanisms that determine
higher-order chromosome structure. The
involvement of bona fide condensin subunits
in dosage compensation together with our
observation that the DCC acts at a distance to
regulate gene expression suggested that the
DCC might alter the topology of X
chromosomes to reduce gene expression
chromosome wide.
Using genome-wide chromosome
conformation capture techniques (in
collaboration with Job Dekker's lab at U. Mass.
Worcester) with single-cell fluorescence in situ
hybridization and RNA-seq to compare
chromosome structure and gene expression
in wild-type and dosage-compensation-
defective embryos, we showed that the DCC
remodels X chromosomes of hermaphrodites
into a unique, sex-specific spatial
conformation, distinct from autosomes, using
its highest-affinity rex sites to facilitate
long-range interactions across X.
Dosage-compensated X chromosomes consist
of self-interacting domains (~ 1 Mb)
resembling mammalian Topologically
Associating Domains (TADs). TADs on X have
stronger boundaries and more regular
spacing than those on autosomes. Many TAD
boundaries on X coincide with the
highest-affinity rex sites, and these boundaries
become diminished or lost in mutants lacking
DCC binding, causing the structure of X to
resemble that of autosomes. These results
predicted that deletion of an endogenous rex
site at a DCC-dependent boundary should
disrupt the boundary. As predicted,
Cas9-mediated deletion of a rex site greatly
diminished the boundary, further
demonstrating the condensin-driven
remodeling of X-chromosome topology during
dosage compensation. Thus, condensin acts
as a key structural element to reorganize
interphase chromosomes and thereby
regulate gene expression. Prior to our work,
no molecular trigger or set of DNA binding
sites was known to cause a comparably strong
effect on TAD structure in higher eukaryotes.
Our understanding of the topology of
dosage-compensated X chromosomes
provides fertile ground to decipher the
detailed mechanistic relationship between
higher-order chromosome structure and
chromosome-wide regulation of gene
expression.
X-Chromosome Domain Architecture
Regulates C. elegans Lifespan but Not
Dosage Compensation.
Interphase chromosomes are organized into a
series of structures ranging from
kilobase-scale chromatin loops to one
megabase-scale topologically associating
domains (TADs) and hundred-megabase
territories. Mechanisms that establish these
higher-order chromosome structures and
their roles in gene regulation have been
elusive.
Understanding the relationship between TAD
structure and gene expression in mammalian
cells has been challenging because
architectural proteins that establish TADs also
bind and function at locations other than TAD
boundaries, such as promoters, making it
unclear whether transcriptional changes
resulting from their depletion are caused by
altered TAD structure or by the proteins' other
roles in gene regulation. Furthermore, the
architectural proteins that establish
mammalian TADs, such as condensin
complexes, also play roles in essential cellular
processes such as chromosome segregation,
making the significance of TADs difficult to
assess at the organismal level by depleting the
proteins.
X chromosome dosage compensation in C.
elegans has been ideal for dissecting the roles
of TADs. Binding of the condensin DCC to X
results in eight DCC-dependent TAD
boundaries. All eight boundaries coincide
with a high-affinity DCC rex site. Without DCC
binding, the eight TAD boundaries are lost,
causing X structure to resemble that of
autosomes with fewer, less regularly spaced
TAD boundaries. These remaining boundaries
on X are DCC independent. Rather than
depleting condensin subunits to disrupt TADs,
we dissected the mechanism of TAD
formation and the function of TADs by
deleting a series of rex sites at TAD
boundaries. We then measured the resulting
chromosome structure and assessed the
effect on gene expression and animal
development. We also inserted high-affinity
rex sites at new locations on 8rexΔ and
wild-type X chromosomes to determine
whether one rex site is sufficient to establish a
new TAD boundary.
Each rex deletion eliminated the associated
DCC-dependent TAD boundary, revealing that
DCC binding at a high-occupancy rex site is
necessary for boundary formation. Insertion
of a rex site at a new location on X defined a
new boundary, indicating that DCC binding at
a high-occupancy rex site is sufficient to define
a boundary on X. Deleting all eight rex sites at
the eight DCC-dependent boundaries
recapitulated the TAD structure of a DCC
mutant. These 8rexΔ animals provided a
unique opportunity to measure transcription
when TAD structure was grossly disrupted
across an entire metazoan chromosome but
binding of the key architectural protein
complex persisted on the numerous
remaining rex sites. The 8rexΔ worms lacked
canonical dosage compensation phenotypes
and had normal compaction of X
chromosomes. Embryos did not show
statistically significant changes in
X-chromosome expression, indicating that
TAD structure does not drive dosage
compensation. The absence of TADs allowed
us to identify additional DCC-mediated
X-chromosome structure: the DCC promotes
DNA interactions across X between loci within
0.1-1 Mb. These TAD-independent
interactions may underlie X compaction and
be important for transcriptional repression.
Although abrogating TAD structure in
hermaphrodites by deleting rex sites did not
disrupt dosage compensation, it did reduce
thermotolerance, accelerate aging, and
shorten lifespan, implicating chromosome
architecture in stress responses and aging.
Targeted Genome-editing Across Highly
Diverged Nematode Species.
Thwarted by the lack of reverse genetic
approaches to enable cross-species
comparisons of gene function, we established
robust strategies for targeted genome editing
across nematode species diverged by 300
MYR. In our initial work, a collaboration with
Sangamo BioSciences, we used engineered
nucleases containing fusions between the
DNA cleavage domain of the enzyme FokI and
a custom-designed DNA binding domain:
either zinc-finger motifs for zinc-finger
nucleases or transcription activator-like
effector domains for TALE nucleases (TALENs).
In those experiments, we allowed the DNA
double-strand breaks to be repaired
imprecisely by non-homologous end joining
(NHEJ) to create mutations in precise locations.
We then extended the use of TALENs to
achieve precise insertion and deletion of
desired sequences by introducing
single-stranded or double-stranded templates
to generate precise insertions or deletions
through homology directed repair (HDR), the
first demonstration of HDR using ZFNs or
TALENs in the nematode community. We then
adopted the use of the CRISPR-associated
nuclease Cas9 because of the ease in making
RNA guides to program target specificity.
Despite successful application of Cas9
technology, predicting DNA targets and guide
RNAs that support efficient genome editing
was problematic. We then devised a strategy
for high-frequency genome editing (both NHEJ
and HDR) at all targets tested. The key
innovation was designing guide RNAs with a
GG motif at the 3' end of their target-specific
sequences. This design increased the
frequency of mutagenesis 10-fold. The ease of
mutant recovery was further enhanced by
combining this efficient guide design with a
co-conversion strategy, in which targets of
interest are analyzed in animals exhibiting a
dominant phenotype caused by Cas9-
dependent editing of an unrelated target.
Evolution cis-acting Regulatory Sites that
Control Dosage Compensation.
Mechanisms that specify sexual fate and
compensate for X-chromosome dose have
diverged rapidly across species compared to
other developmental processes, making it
particularly informative to study these rapidly
changing processes over short evolutionary
time scales. Application of our genome editing
strategies to C. briggsae revealed that the core
dosage compensation machinery and key
components of the genetic hierarchy that
controls dosage compensation and sex
determination were conserved across the 30
MYR separation between C. elegans and C.
briggsae. In contrast, the set of cis-acting
elements on X that recruit the DCC (rex sites)
has diverged, retaining no functional overlap.
ChIP-seq analysis defined the C. briggsae DCC
binding sites, and in vivo binding assays
confirmed the ability of these sites to recruit
the DCC when detached from X in C. briggsae
but not in C. elegans, and vice versa. The
evolution of these sites differs dramatically
from the highly conserved DCC binding sites
used by equivalently diverged fruit fly species
and from the unchanged target sites of
conserved transcription factors that control
multiple developmental processes from flies
to humans. Hence, the divergence in DCC
binding specificity across nematode species
provides a powerful opportunity to
understand the path and timing for the
concerted change in hundreds of DNA target
sites and the evolution of X chromosomes. We
have extended our analysis of DCC binding
specificity to other nematode species and
have shown that rex sites have diverged
functionally at least three times in 30 MYR of
evolutionary history.
Tethering Replicated Chromosomes via
Cohesin to Ensure Genome Stability during
Meiosis.
Faithful segregation of chromosomes during
cell division is essential for genome stability.
Accurate chromosome segregation is required
both for the proliferative cell divisions that
produce daughter cells during mitosis and the
two sequential divisions that produce haploid
sperm and eggs from diploid germline stem
cells during meiosis. Approximately 30% of
human zygotes have abnormal chromosome
content at conception due to defects in
meiosis. Such aneuploidy is a leading cause of
miscarriages and birth defects and arises, in
part, from defects in sister chromatid
cohesion (SCC). SCC tethers replicated sister
chromatids prior to cell divisions to ensure
proper chromosome segregation. In humans,
SCC is established in the developing germ cells
of a fetus and must be maintained until
ovulation in adults. This long-lived SCC is
established and maintained by cohesin
complexes, evolutionarily conserved protein
complexes structurally related to condensin
(Figure 7).
Studies in budding yeast showed that mitotic
and meiotic cohesins are distinct but differ
only in a single subunit called the kleisin.
During yeast meiosis, a single cohesin
complex carries out all aspects of SCC. In
contrast, our work in nematodes shows that
regulation of meiotic SCC in higher eukaryotes
is more complex. We found that multiple
functionally specialized cohesin complexes
mediate the establishment and two-step
release of SCC that underlies the production
of haploid gametes (Figure 7). The meiotic
complexes differ by a single kleisin subunit,
and the kleisin influences nearly all aspects of
meiotic cohesin function: the mechanisms for
loading cohesins onto chromosomes, for
triggering DNA-bound cohesins to become
cohesive, and for releasing cohesins in a
temporal- and location-specific manner
(Figure 8). One kleisin triggers cohesion just
after the chromosomes replicate, as in yeast.
Unexpectedly, the other triggers cohesion in a
replication-independent manner, only after
programmed DSBs are made during meiosis
to initiate recombination between
homologous maternal and paternal
chromosomes. Thus, break-induced cohesion
is essential for tethering replicated meiotic
chromosomes. Later, recombination
stimulates separase-independent removal of
the two different cohesin complexes from
reciprocal chromosomal territories flanking
the crossover site. This region-specific
removal likely underlies the two-step
separation of homologs and sisters.
Unexpectedly, one cohesin complex also
performs cohesion-independent functions in
synaptonemal complex assembly. Our
findings establish a new model for cohesin
function in meiosis: the choreographed
actions of multiple cohesins, endowed with
unexpectedly specialized functions by their
kleisins, underlie the stepwise separation of
homologous chromosomes and then sister
chromatids required for reduction of genome
copy number. This model diverges
significantly from that in yeast but likely
applies to plants and mammals, which utilize
similar meiotic kleisins.
Meiotic Chromosome Structure Constrain
and Respond to Designation of Crossover
Sites.
Crossover recombination events between
homologous chromosomes are required to
form chiasmata, temporary connections
between homologues that ensure their proper
segregation at meiosis I. Despite this
requirement for crossovers and an excess of
the double-strand DNA breaks that are the
initiating events for meiotic recombination,
most organisms make very few crossovers per
chromosome pair. Moreover, crossovers tend
to inhibit the formation of other crossovers
nearby on the same chromosome pair, a
poorly understood phenomenon known as
crossover interference. We showed (in
collaboration with the Villenueve lab at
Stanford) that the synaptonemal complex, a
meiosis-specific structure that assembles
between aligned homologous chromosomes,
both constrains and is altered by crossover
recombination events. Partial depletion of the
synaptonemal complex central region
proteins attenuates crossover interference,
increasing crossovers and reducing the
effective distance over which interference
operates, indicating that synaptonemal
complex proteins limit crossovers. Moreover,
we showed that crossovers are associated
with a local 0.4-0.5-micrometre increase in
chromosome axis length. We proposed that
meiotic crossover regulation operates as a
self-limiting system in which meiotic
chromosome structures establish an
environment that promotes crossover
formation, which in turn alters chromosome
structure to inhibit other crossovers at
additional sites.
Ironically, the effect of depleting condensin I
or condensin II on increasing crossovers
appears to occur by a different mechanism,
because the sites of extra crossovers are not
marked by the same molecular markers as
the crossovers created by reducing the
synaptonemal complex. We are investigating
the crossover pathway employed to achieve
these extra, non-interfering crossovers in
condensin mutants.