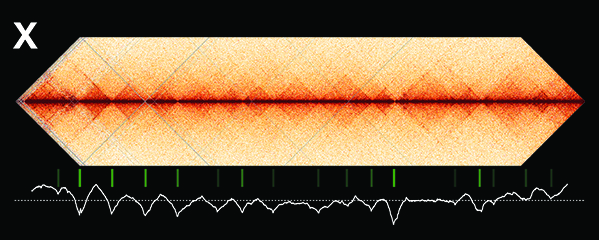
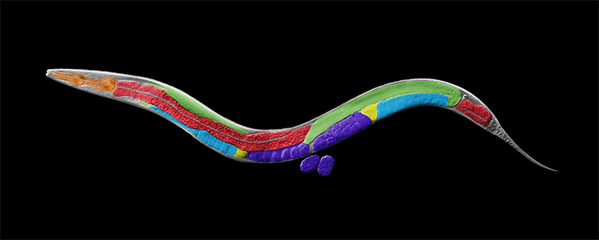
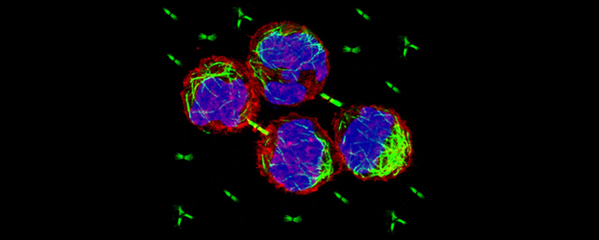
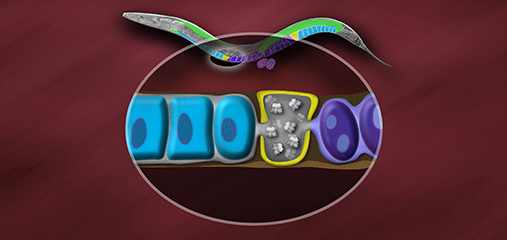
Our research focuses on the interplay between the structure of chromosomes and their function. Chromosomes undergo dynamic behaviors during development to ensure genome stability and accurate cell fate decisions. We study inter-related molecular networks that control diverse chromosome behaviors: chromosome counting to determine sexual fate; X-chromosome remodeling to achieve X-chromosome repression during dosage compensation, an epigenetic process; chromosome cohesion to tether and release replicated chromosomes for reducing genome copy number during germ cell formation; and chromosome compaction to control gene expression, chromosome segregation, and recombination between maternal and paternal chromosomes. We have found that the developmental control of gene expression is achieved through chromatin modifications that affect chromosome structure with epigenetic consequences. We have also established robust procedures for targeted genome editing across nematode species diverged by 300 MYR to study the evolution of sex determination and dosage compensation. We combine genetic, genomic, proteomic, biochemical, and cell biological approaches to study these questions in the model organism Caenorhabditis elegans, a round worm, and its related nematode species.
Our lab is affiliated with Howard Hughes Medical Institute and UC Berkeley.
We are in the Department of Molecular and Cell Biology.