Current Projects
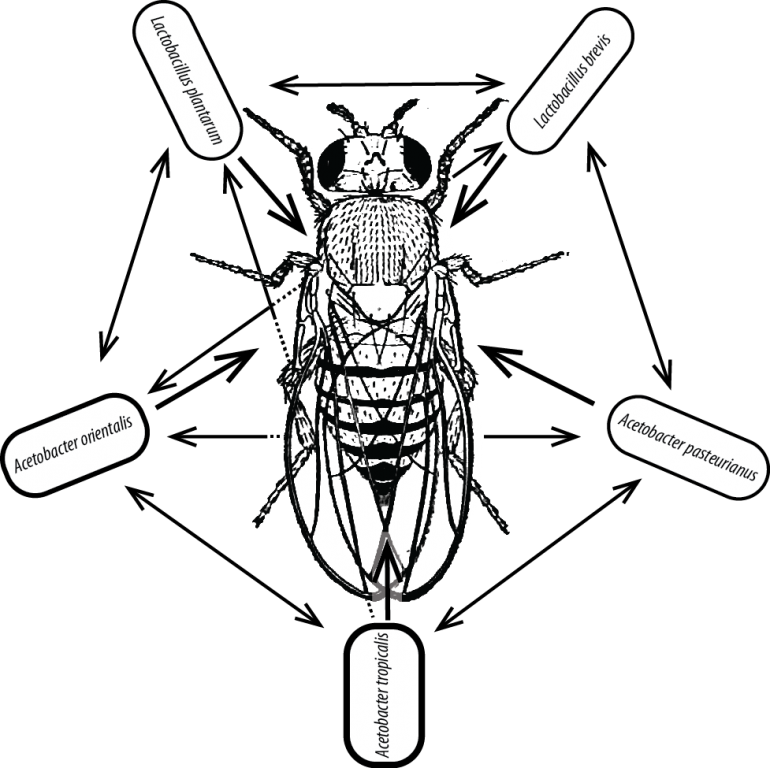
Our lab primarily focuses on a model microbiome in the fruit fly Drosophila melanogaster. The fly gut microbiome is a natural and complete with as few as 5 species of bacteria. We use this reductionist system to study host-microbiome interactions as well as basic principles of ecology.
1. Gut microbial ecology modulates fly health. Using gnotobiotic flies and a battery of physiological and fitness assays, we study the effects of the gut community on fly health. Angus Chandler and Vivian Zhang lead projects in this area.
In collaboration with Carolyn Elya and Mike Eisen (UC Berkeley), we are examining microbiota-induced alterations to fly gene expression.
2. Community ecology of the gut microbiota. Using both gnotobiotic flies and an in vitro system with high throughput growth assays, we study the microbial interactions that shape the fly gut community. Tuzun Guvener leads this project. We collaborate with Michi Taga (UC Berkeley) to study the specific interaction mechanisms. We collaborate with Andres Aranda-Diaz and KC Huang (Stanford) to measure interactions at the single cell level. We collaborate with Hans Carlson and Adam Deutschbauer (LBNL) to make high throughput measurements.
3. Microbial population dynamics in single hosts and in host populations. We have developed single fly feeding and monitoring techniques to measure microbial populations in the gut. Using these approaches, we are studying basic principles of colonization and stability in the context of community interactions and disease ecology. Benjamin Obadia leads this project.
Collaborations:
Phage-based enterotherapies. In collaboration with Richard Calendar (UC Berkeley) and Michi Taga (UC Berkeley), we are developing phage-based enterotherapies to push ecological state change in the gut microbiome.
Groundwater metagenomics in agriculture-impacted regions. In collaboration with Thomas Harter, Rob Atwill and Xunde Li (UC Davis), Clarissa Nobile and Aaron Hernday (UC Merced), Glen Wallace (Pacific Groundwater Group), and Joe DeRisi (UCSF), we use shotgun metagenomics to discover the microbial genomic basis of agricultural contamination impacts on groundwater and develop remediation strategies.
Outbreeding the Norwegian Lundehund to reduce persistent autoimmune disease. Through the Sather Center (UC Berkeley) and in collaboration with Claudia Melis (Norwegian University of Science and Technology) and Ottmar Distl (Institut für Tierzucht, Hannover), we study the genomic and gut metagenomic basis for a highly prevalent and lethal autoimmune disease in this charismatic, puffin-hunter dog breed.
Funding:
The lab is funded by the NIH Director's Early Independence Award Program and the Berkeley MCB Department's Bowes Fellows Program.