
Current projects
The frz genes control movement in M. xanthus
Reversal frequency is essential for the correct functioning of both A and S motility as well as for development. 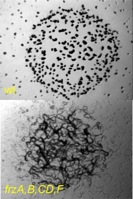 The reversal frequency is regulated by a sensory transduction pathway composed of several factors encoded by the frz operon. frz genes were cloned and sequenced by our lab and the annotation “frz” derives from the “frizzy” phenotype (defect in fruiting body formation see figure to the right, lower panel) observed on developmental media in strains carrying deletions in the frz operon.
The reversal frequency is regulated by a sensory transduction pathway composed of several factors encoded by the frz operon. frz genes were cloned and sequenced by our lab and the annotation “frz” derives from the “frizzy” phenotype (defect in fruiting body formation see figure to the right, lower panel) observed on developmental media in strains carrying deletions in the frz operon.  The frz genes are homologous to chemotaxis genes of enteric bacteria. Despite the similarities between the chemotaxis proteins of enteric bacteria and M. xanthus Frz proteins, fundamental differences exist between these different bacteria in the ability of cells to recognize and respond to substances in their environment. From our analysis of the gene products, we have constructed a model of the frz sensory transduction pathway. In our model the Frz core receive inputs through the methyl accepting chemotaxis protein FrzCD. We are interested in studying the mechanism by which FrzCD is stimulated since it lacks transmembrane and periplasmic domains normally involved in the sensing of external signals.
The frz genes are homologous to chemotaxis genes of enteric bacteria. Despite the similarities between the chemotaxis proteins of enteric bacteria and M. xanthus Frz proteins, fundamental differences exist between these different bacteria in the ability of cells to recognize and respond to substances in their environment. From our analysis of the gene products, we have constructed a model of the frz sensory transduction pathway. In our model the Frz core receive inputs through the methyl accepting chemotaxis protein FrzCD. We are interested in studying the mechanism by which FrzCD is stimulated since it lacks transmembrane and periplasmic domains normally involved in the sensing of external signals.
 The reversal frequency is regulated by a sensory transduction pathway composed of several factors encoded by the frz operon. frz genes were cloned and sequenced by our lab and the annotation “frz” derives from the “frizzy” phenotype (defect in fruiting body formation see figure to the right, lower panel) observed on developmental media in strains carrying deletions in the frz operon.
The reversal frequency is regulated by a sensory transduction pathway composed of several factors encoded by the frz operon. frz genes were cloned and sequenced by our lab and the annotation “frz” derives from the “frizzy” phenotype (defect in fruiting body formation see figure to the right, lower panel) observed on developmental media in strains carrying deletions in the frz operon.  The frz genes are homologous to chemotaxis genes of enteric bacteria. Despite the similarities between the chemotaxis proteins of enteric bacteria and M. xanthus Frz proteins, fundamental differences exist between these different bacteria in the ability of cells to recognize and respond to substances in their environment. From our analysis of the gene products, we have constructed a model of the frz sensory transduction pathway. In our model the Frz core receive inputs through the methyl accepting chemotaxis protein FrzCD. We are interested in studying the mechanism by which FrzCD is stimulated since it lacks transmembrane and periplasmic domains normally involved in the sensing of external signals.
The frz genes are homologous to chemotaxis genes of enteric bacteria. Despite the similarities between the chemotaxis proteins of enteric bacteria and M. xanthus Frz proteins, fundamental differences exist between these different bacteria in the ability of cells to recognize and respond to substances in their environment. From our analysis of the gene products, we have constructed a model of the frz sensory transduction pathway. In our model the Frz core receive inputs through the methyl accepting chemotaxis protein FrzCD. We are interested in studying the mechanism by which FrzCD is stimulated since it lacks transmembrane and periplasmic domains normally involved in the sensing of external signals. FrzCD activity is regulated by its methylation level. A modulation of FrzCD methylation, during development, occurs at the level of a specific domain (TPR) in the methyltransferase FrzF, as well as the level of the histidine kinase FrzE. The identification of FrzCD methylation sites is under investigation. Once activated, FrzCD is able to activate the histidine kinase FrzE to autophosphorylate (see figure on the left). The downstream component FrzZ, a CheY-CheY fusion, accepts phosphate from FrzE. Regulation of FrzE autophosphorylation and subsequent signaling events that ultimately trigger a reversal in M. xanthus, is currently under investigation.
Characterization of the social motility protein FrzS
We are currently examining the role of the protein FrzS in type IV pili (TFP)-dependent motility. frzS mutants are completely defective in colony-level TFP-powered movement (also known as social motility in M. xanthus). The FrzS protein localizes to the cell poles and may play a role in the proper function of the polar TFP during movement in large groups of cells. Furthermore, FrzS is specifically enriched at the leading cell pole as cells move forward and, upon cell reversal, FrzS switches and becomes enriched at the new leading pole (see FrzS-GFP movie on the right). We are currently identifying in cis and in trans regulators of FrzS subcellular localization. Recently, we have found that several protein domains of FrzS are required for its normal dynamic pattern of subcellular localization and that mislocalization of FrzS in the cell leads to severe defects in M. xanthus social motility.
AglZ is part of focal adhesion complexes involved in A motility.
AglZ is a protein essential for adventurous motility (the motility that doesn’t require type IV pilus), but dispensable for social motility. In motile cells, the protein is localized in cluster spanning the cell length. As cells move forward, AglZ clusters maintained fixed positions with respect to the substrate and the periodicity of AglZ clusters is very similar to the helical pitch of bacterial actin MreB. Because AglZ clusters remain fix relatively to the substratum as cells move forward they must be moving in the opposite direction to that of the cell. These dynamics suggest the formation of cytoskeleton-anchored transient surface adhesions that could power cellular movement.
 A cartoon depiction of the two M. xanthus motility systems. Social motility is mediated by the extension and retraction of type IV pili (black tendrils) at the leading pole of the cells. Adventurous motility involves multiple transient adhesion complexes (colored ovals on the substratum of cells) located throughout the length of the cell.
A cartoon depiction of the two M. xanthus motility systems. Social motility is mediated by the extension and retraction of type IV pili (black tendrils) at the leading pole of the cells. Adventurous motility involves multiple transient adhesion complexes (colored ovals on the substratum of cells) located throughout the length of the cell.