Megan Martik

Assistant Professor of Genetics, Genomics, Evolution, and Development
Research Interests
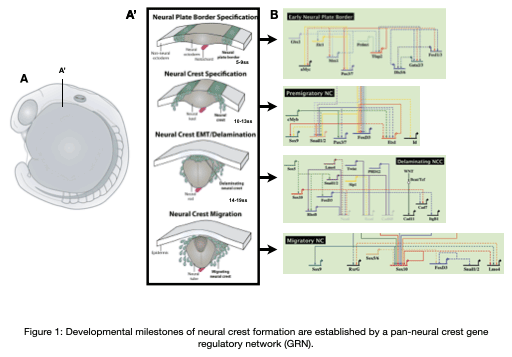
The neural crest is an important stem cell population in the embryo characterized by its multipotency, migratory behavior, and broad ability to differentiate into derivatives as diverse as cardiomyocytes, craniofacial skeleton, and the peripheral nervous system. Underlying the development of these unique derivatives is a neural crest gene regulatory network (GRN) that describes the regulatory interactions at each stage of neural crest development (Figure 1). Our team is focused on understanding the regulatory networks controlling the development of the neural crest from a multipotent stem cell population into unique derivatives, how these networks are re-used during adult repair processes, how these networks become dysregulated at the onset of disease, and how these networks evolve to give rise to morphological novelties.
Current Projects
Project 1: Gene network control dictating the differentiation of the cardiac neural crest
Cardiac neural crest cells migrate through the developing embryo to contribute to important portions of the cardiovascular system by contributing to cardiomyocytes, coronary vessels, the outflow tract, valves, and parasympathetic innervation (Movie 1). Dysregulation and abnormalities in the developmental programs of the cardiac neural crest result in a broad range of human birth defects, including persistent truncus arteriosus, abnormal myocardium function, and misalignment of the arch arteries. Furthermore, in adult zebrafish, it has been shown that ablation of neural crest-derived cardiomyocytes results in severe hypertrophic cardiomyopathy, altered cardiomyocyte size, diminished heart capacity, and heart failure during cardiac stress tests. Using zebrafish as a model, our goal is to understand how GRNs coordinate differentiation of the cardiac neural crest into unique cardiac derivatives and how these developmental programs can go awry and result in improper heart function and congenital heart defects.
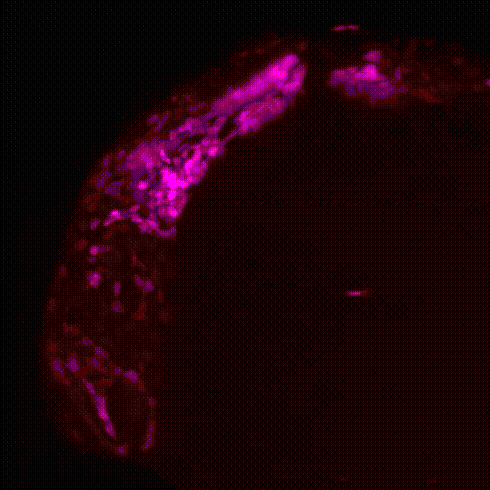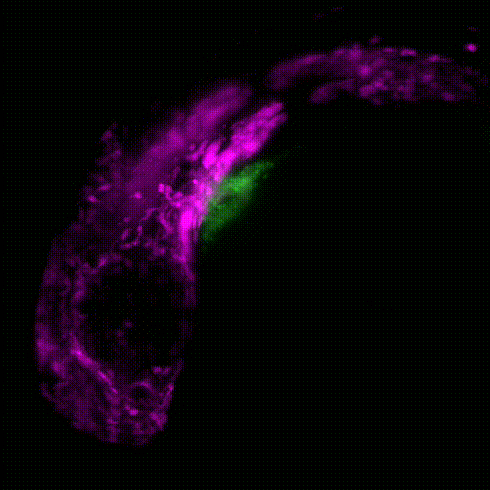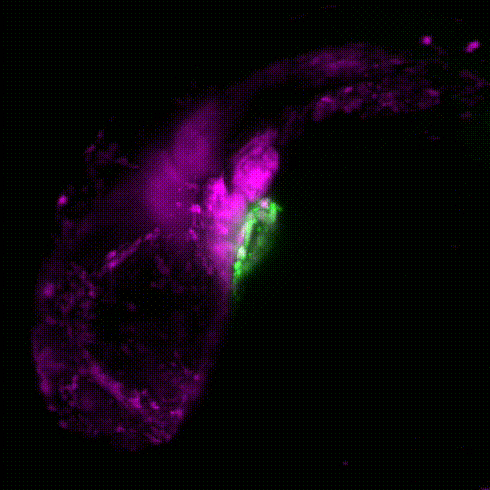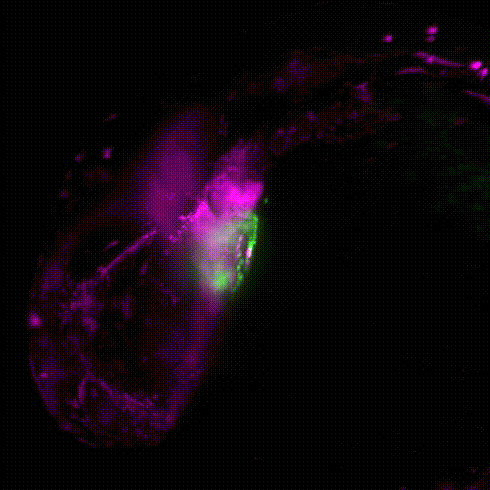
Movie 1: Transgenic reporter lines and light sheet microscopy are used to track the migration of neural crest cells (magenta) to the developing zebrafish heart (green).
Project 2: Neural crest contributions to adult tissue regeneration
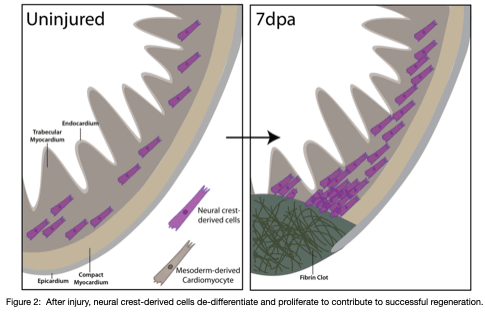
Our recent work reveals a previously unrecognized contribution of neural crest-derived cells to cardiomyocytes across vertebrates and their role during regeneration in the adult heart of zebrafish (Tang*, Martik*, et al, eLife, 2019) (Figure 2). We are interested in how neural crest-derived cells and redeployment of an embryonic-like gene regulatory program control the de-differentiation and proliferation, fibrosis, and scar formation/regression during the cardiac regeneration process in zebrafish. Further, by uncovering GRN differences between amniote hearts, which lack the capacity to regenerate, and anamniotes with remarkable regenerative ability, our goal is to drive regeneration in systems where it otherwise fails to occur.
Project 3: Neural crest in vertebrate evolution
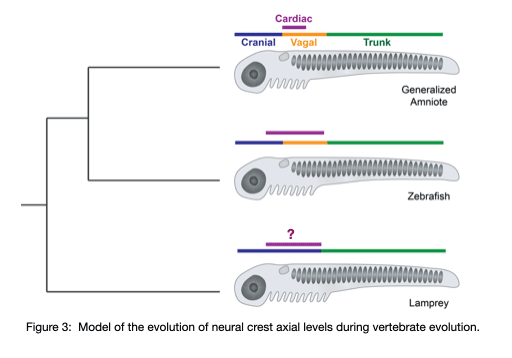
Using the jawless fish, the sea lamprey, our lab group is working to understand the gene regulatory changes that drove the diversification of the neural crest and the evolution of vertebrates. Our previous work (Martik, et al, Nature 2019) found that the lamprey neural crest gene network is missing crucial information that is required for axial refinement of the neural crest. Throughout gnathostome evolution, the neural crest gene network was progressively elaborated to modulate differentiation capacity along the anterior-posterior axis. Our work aims to uncover the progressive assembly of a novel axial-specific regulatory circuits that allowed for the elaboration of the neural crest during vertebrate evolution with emphasis on cardiac neural crest derivatives (Figure 3).
Project 4: Dysregulation of neural crest GRNs in neural crest-derived cancers
Neuroblastomas emerge at sites along the sympathetic nervous system as a result of improper differentiation of neural crest-derived sympathoadrenal progenitors; however, the molecular mechanisms underlying the ontogeny of neuroblastoma are not fully understood. Determining the gene regulatory network circuitry underlying sympathoadrenal development and comparing this to that active at the onset of neuroblastoma holds the promise of yielding insights into new therapeutic genetic targets. Using the zebrafish as a model, our group is working to unravel the gene regulatory network controlling the differentiation of the sympathetic nervous system with the long term goal of providing information about potential therapies for prevention and treatment.
For more information: www.martiklab.org
Selected Publications
For up-to-date publications from the Martik lab: https://bit.ly/3ySrFYJ
Martik ML and Bronner ME. (2021) Riding the crest to get a head: neural crest evolution in vertebrates. Nature Reviews Neuroscience. (in press)
Martik ML. (2020) Metabolism makes and mends the heart. eLife. 2020;9:e54665 (doi: 10.7554/eLife.54665)
McClay DR, Warner JF, Martik ML, Miranda E, Slota L. (2020) Gastrulation in the sea urchin. Current Topics in Developmental Biology. 2020;136:195-218.(https://doi.org/10.1016/bs.ctdb.2019.08.004)
Martik ML, Gandhi S, Uy BR, Green SA, Gillis JA, Simoes-Costa M, and Bronner ME. (2019) Evolution of the New Head by gradual acquisition of neural crest regulatory circuits. Nature. 220, 268-4.
Tang W*, Martik ML*, Li Y, and Bronner ME. (2019) Cardiac neural crest contributes to cardiomyocytes in amniotes and heart regeneration in zebrafish. eLife. 2019;8:e47929 (doi:10.7554/eLife.47929)
Martik ML and Bronner ME. (2017) Regulatory logic underlying diversification of the neural crest. Trends in Genetics. 33(10):715-727. (http://dx.doi.org/10.1016/j.tig.2017.07.015)
Martik ML and McClay DR. (2017) New insights from a high-resolution look at gastrulation in the sea urchin, Lytechinus variegatus. Mechanisms of Development. pii: S0925-4773(17)30002-3. (doi: 10.1016/j.mod.2017.06.005)
Martik ML, Lyons DC, McClay DR. (2016) Developmental gene regulatory networks and what we can learn from them. F1000Research, 5(F1000 Faculty Rev):203 (doi: 10.12688/f1000research.7381.1).
Israel JW, Martik ML, Byrne M, Raff EC, Raff RA, McClay DR, Wray GA. (2016) Comparative Developmental Transcriptomics Reveals Rewiring of a Highly Conserved Gene Regulatory Network during a Major Life History Switch in the Sea Urchin Genus Heliocidaris. PLoSBiology. 14(3): e1002391. (doi:10.1371/journal.pbio.1002391.)
Martik ML and McClay DR. (2015). Deployment of a retinal determination gene network drives directed cell migration. eLife. 2015;10.7554/eLife.08827.
Lyons DC, Martik ML, Saunders LR, McClay DR. (2014). Specification to biomineralization: following a single cell type as it constructs a skeleton. Integrative and Comparative Biology. 54(4):723-733.
McIntyre DC, Lyons DC, Martik ML, McClay DR. (2014.) Branching out: origins of the sea urchin larval skeleton in development and evolution. Genesis. 52(3):173-185.
Martik ML and McClay DR. (2013). Gut development in the sea urchin.Translational Gastroenterology, from Development to Disease. John Wiley & Sons, Inc: Hoboken, New Jersey
Yankura KA, Martik ML, Jennings CK, Hinman VF. (2010). Uncoupling of complex regulatory patterning during evolution of larval development in echinoderms. BMC Biology. Nov 30;8:143.
*equal contribution
Last Updated 2021-08-05