Lin He

Thomas and Stacey Siebel Distinguished Chair in Stem Cell Research and Professor of Cell Biology, Development and Physiology*
*And Affiliate, Division of Immunology & Pathogenesis
Research Interests
In comparative genomic studies, the number of protein-coding genes clearly fails to correlate with the developmental and pathological complexity in mammals. Our overall research interest is to understand the unique biological functions and molecular regulation of various non-coding RNAs in development and disease. My postdoctoral work has identified some of the first miRNAs in key oncogene and tumor suppressor pathways. Studies from my own lab has extended beyond mere identification of functionally important miRNAs; we aim to understand the distinct biological functions and molecular regulation conferred by miRNAs and other non-coding elements in development and disease using an interdisciplinary approach combining mouse genetics, genomics, Xenopus genetics, cell biology, and molecular biology. Our studies have provided important insights on the fundamental molecular mechanisms that govern the unique functional complexity of the non-coding genome.
Current Projects
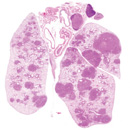 miRNAs in cancer biology
miRNAs in cancer biology
Non-coding RNAs, particularly microRNAs (miRNAs) are increasingly recognized as integral components of oncogene and tumor suppressor network. Using mouse models for lung cancer, lymphoma and leukemia, we aim to characterize the functional importance of multiple miRNAs in tumor progression and tumor metastasis. We are particularly interested in understanding the unique structural and function importance of non-coding RNAs in cancer development.
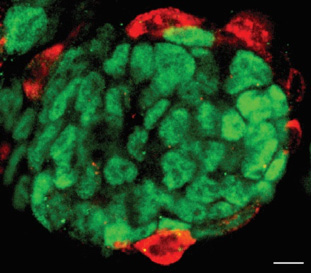 non-coding RNAs in stem cell biology
non-coding RNAs in stem cell biology
Mouse embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) exhibit pluripotent potential, contributing largely to embryonic lineages, but not extra-embryonic, lineages. Yet this restricted developmental potential could be expanded upon alteration of specific non-coding RNAs. Thus, a network of proteins, ncRNAs and retrotransposons constitutes the molecular basis that defines the pluripotent cell fate potential.
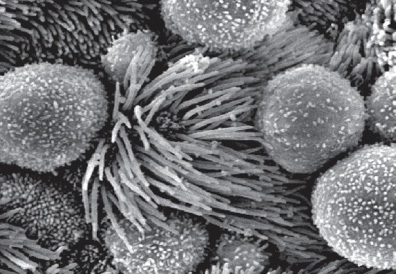 ncRNAs and retrotransposons in development
ncRNAs and retrotransposons in development
Functional importance of the non-coding genome remains largely unexplored. Using mouse genetics and Xenopus genetics, we have demonstrated the importance of multiple conserved non-coding RNAs in diverse developmental processes, including preimplantation development, ciliogenesis, and lung development. Thus, an intricate molecular network of proteins and non-coding RNAs play an essential role in development and disease.

CRISPR genome editing in mice
CRISPR-mediated mouse genome editing is typically accomplished by microinjection of Cas9 DNA/RNA and sgRNA into zygotes to achieve editing in one step. We developed a simple and economic electroporation-based strategy, designated as CRISPR RNP Electroporation of Zygotes (CRISPR-EZ), to deliver Cas9/sgRNA ribonucleoproteins (RNPs) into mouse zygotes with 100% efficiency. CRISPR-EZ enables highly efficient and high-throughput genome editing in vivo, with a significant improvement in embryo viability compared with microinjection-based technology
.
Selected Publications
Chen S, Lee B, Lee A, Modzelewski A and He L. Highly Efficient Mouse Genome Editing by CRISPR Ribonucleoprotein Electroporation of Zygotes. J. Biol Chem 2016 May 5. pii: jbc.M116.733154.
Olive V, Sabio E, Minella AC and He L. Outside the coding genome, microRNAs make a big difference. Science Signaling, Sci Signal. 2015 Mar 17;8(368):re2.
Song R*, Walentek P*, Sponer N*, Lize M, Klimke A, Lee JS, Dixon GR, Lishko P, Kessel M, He L. miR-34/449 miRNAs are required for motile ciliogenesis by repressing Cp110. Nature, research article, 2014 Jun 5;510(7503):115-20.
Krzeszinski JY, Wei W, Huynh H, Jin Z, Chang T, Xie X, He L, Lingegowda MS, Lopez-Berestein G, Sood AK, Mendell JT and Wan Y. MiR-34a blocks Osteoporosis and Bone Metastasis by Inhibiting Osteoclastogenesis and Tgif2. Nature, 2014 Aug 28;512(7515):431-5.
Xue B and He L. An expanding universe of the non-coding genome in cancer biology. Carcinogenesis, 2014 Jun;35(6):1209-1216. Epub 2014 Apr 18.
Okada N, Ribeiro M, Biton A, Lin C, Lai G, Bu P, Vogel H, Jablons DM , Keller A, Wilkinson JE, He B, Speed TP and He L. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression Genes and Development, 2014 Mar 1;28(5):438-50. doi: 10.1101/gad.233585.113.
Olive V*, Sabio E*, Bennett M, De Jong C, Biton A, McGann JC, Greaney SK, Sodir NM, Zhou AY, Balakrishnan A, Foth M, Luftig M, Goga A, Speed TP, Xuan Z, Evan G, Wan Y, Minella A, He L. A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis. Elife. 2013 Oct 15;2:e00822. doi: 10.7554/eLife.00822. PMID: 24137534
Choi Y*, Lin C*, Zhong Y, He X, Kim S, Okada N, Bu P, Bennett MJ, Chen C, Ozturk A, Hicks G, Hannon GJ, and He L. miR-34a miRNA provides a barrier for somatic cell reprogramming. Nature Cell Biology, 2011 Oct 23;13(11):1353-60. PMID: 22020437.
He L. Post-transcriptional regulation of PTEN dosage by non-coding RNAs. Science Signaling 2010 Nov 2;3(146):pe39. PMID: 21045203.
Olive V *, Bennett MJ*, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li Q, Lowe SW, Hannon GJ# and He L#. miR-19 is a key oncogenic component of mir-17-92. Genes and Development 2009 Dec 15;23(24):2839-49. PMID: 20008935.
He L, He X, Lowe SW and Hannon GJ. microRNAs join the p53 pathway, another piece of the tumor suppressor puzzle. Nat Rev Cancer. 2007 Nov;7(11):819-22.
He L*, He X*, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA# and Hannon GJ#. A microRNA component of the p53 tumour suppressor network. Nature. 2007 Jun 28;447(7148):1130-4.
He L*, Thomson JM*, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ# and Hammond SM#. A microRNA polycistron as a potential human oncogene. Nature. 2005 Jun9; 435(7043):828-33.
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004 Jul;5(7):522-31.
He L, Lu XY, Jolly AF, Eldridge AG, Watson SJ, Jackson PK, Barsh GS# and Gunn TM#. Spongiform encephalopathy caused by defects of an E3 ubiquitin ligase in mahoganoid mice. Science, 2003 Jan 31;299(5607):710-2.
He L*, Gunn TM*, Bouley DM, Lu XY, Watson SJ, Schlossman SF, Duke-Cohan JS, Barsh GS. A biochemical function for attractin in agouti-induced pigmentation and obesity. Nat Genet. 2001 Jan;27(1):40-7.
Gunn TM, Miller KA, He L, Hyman RW, Davis RW, Azarani A, Schlossman SF, Duke-Cohan JS, Barsh GS. The mouse mahogany locus encodes a transmembrane form of human attractin. Nature 1999 Mar 11;398(6723):152-6.
Last Updated 2016-08-16